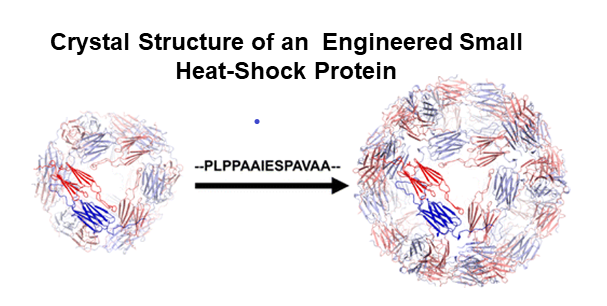
Methodology
The research theme of the lab is to illuminate how protein dynamics – i.e. the twisting, bending, and deformation of the three-dimensional structure, mediate biological function. Specifically, we are interested in understanding how protein kinase function is regulated in signaling pathways, how transporters extrude drugs and various substrates out of the cytoplasm, how transporters reuptake neurotransmitters and solutes, and how molecular chaperones recognize their unfolded protein clients. We also develop methodologies and approaches to address the relationship between structure, function, and dynamics.
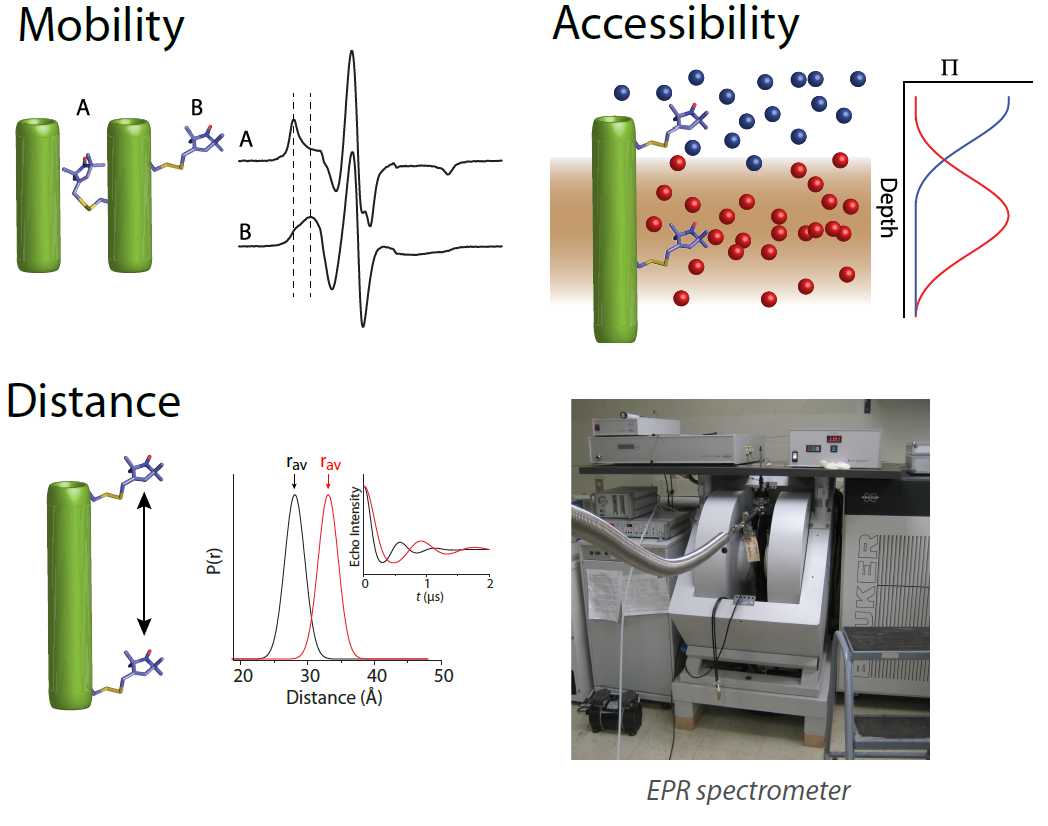
We use a spectrum of tools ranging from biochemical methods, x-ray crystallography, cryo-EM in the test tube to physiological methods in model organisms such as zebrafish. However, our signature methodology, site-directed spin labeling (SDSL) with electron paramagnetic resonance (EPR) spectroscopy can reveal the environment surrounding a spin label and measure the distance between labels. Spin label mobility and accessibility can be measured to reveal information about burial and orientation, and double electron electron resonance (DEER) spectroscopy can measure distances between two spin labels.
We use other methods such as crystallography and cryoEM in collaborative efforts with colleagues at Vanderbilt and other institutions.