Dynamics are Us
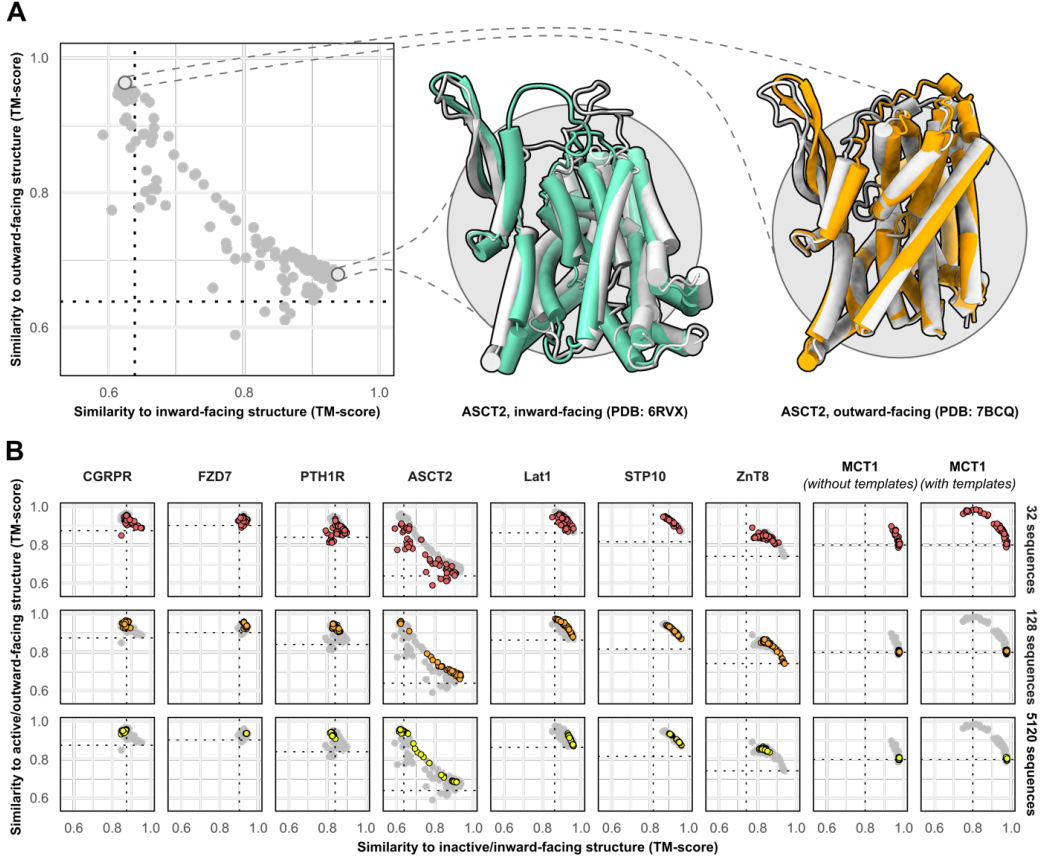
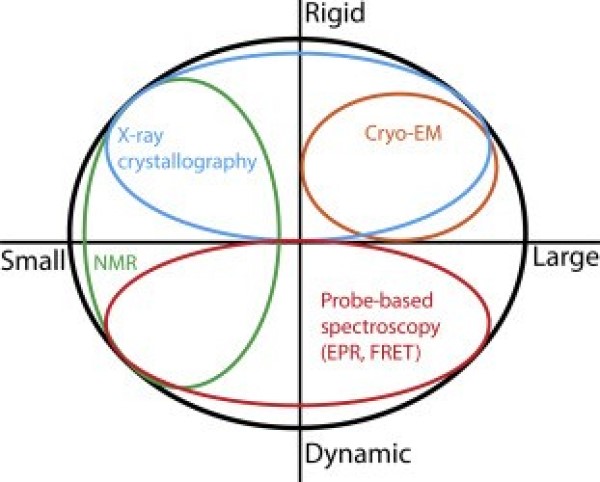
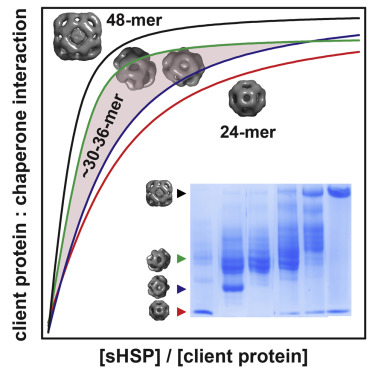
Dynamics represent the fourth dimension linking protein structures to mechanisms. Proteins have parts that gate, bend, twist or catalyze a given reaction. My research program encompasses a wide spectrum of questions that have in common a fundamental interest in how protein dynamics define or regulate biological function. The questions we pursue range from how CaMKII, a critical enzyme in long term memory, decodes the amplitude and frequency of calcium signals; how neurotransmitter transporters couple ion movement to vectorial substrate translocation; how multidrug resistance transporters harness the energy of ATP and transduce it to the mechanical work of substrate extrusion, and how small heat shock proteins, a class of oligomeric molecular chaperones, deploy subunits to recognize and bind unfolding proteins. At the origin of this expansive list of targets is the goal to discover common principles of dynamics. For instance, we have been investigating 12 transporters with the goal of illuminating two central aspects of active transport: namely how these molecules harness various forms of energy and then transduce this energy to drive protein conformational motion.
Why what we study matters?
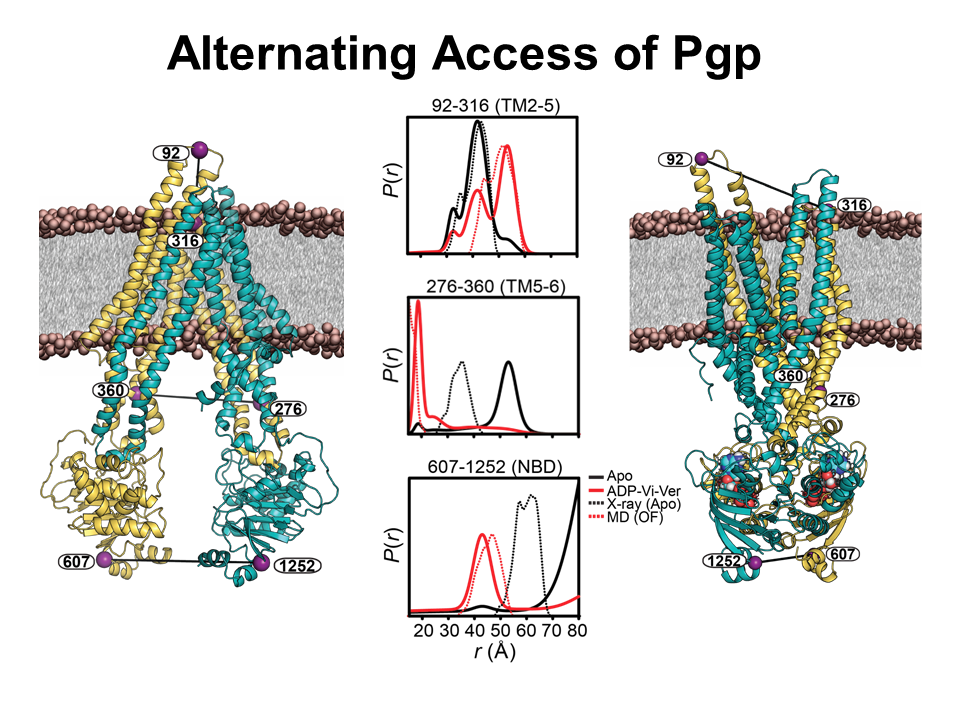
Our systems are selected because of their direct relevance to biomedicine. For example, the major membrane protein involved in drug clearance in humans is Pgp, an ABC transporter involved in resistance to chemotherapy. Thus by understanding how Pgp use the energy of ATP to transport substrates, we can devise approaches for inhibiting this pump and making chemotherapy more effective. Similarly, biogenic amines transporters are the primary site of action psychostimulants and the target of antidepressant. Therefore, an understanding of their functional mechanism is critical in addiction and treatment of depression.
From test tube to model organism
We are currently utilizing a broad range of approaches to integrate biochemical and biophysical perspective in the test tube with whole animal physiology using zebrafish. I am trained as physicist/biophysicist but my research involves diverse approaches in Biochemistry and Cell Biology. Our work on molecular chaperones has recently evolved from a historical focus on biophysical studies in the test tube to testing hypotheses in-vivo using knockout and transgenic strategies in the zebrafish. This new direction is taking on a definitely more cell biology and physiology perspectives. For example, in unpublished work we have pursued a knockout strain that lead to a remarkable heart edema phenotype that is revealing new roles for the protein target in mediating a response to glucocorticoid stress.
In what appears to be a full circle, we have recently began using zebrafish in our transporters studies. Here the interest is to use zebrafish as a model for disease-causing mutations complementing the in-vitro understanding of the effects of these mutations on the conformational cycle. Current work is focused on developing behavioral and physiological assays in knockout strains of ABC transporters linked to human diseases.