McLean Research Group Projects
Current Projects
Copper Complexation Strategies for Differentiation Amino Acid Enantiomers by Drift Tube Ion MobilityInvestigator: Emanuel Zlibut |
 |
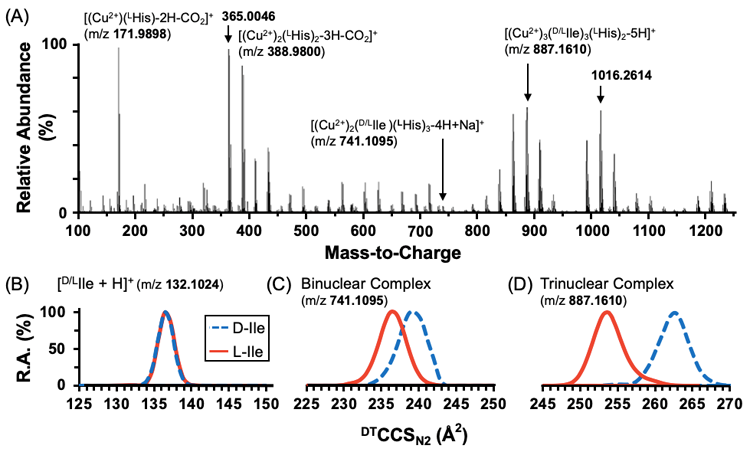
The pursuit of faster, more sensitive analytical techniques for quantitative chiral measurements has led to the development of mass spectrometry-based approaches. In this study, we investigate the separation of chiral molecules forming novel tri-nuclear copper complexes in the presence of a chiral selector that can be directly resolved by ion mobility-mass spectrometry (IM-MS) analyses.
Amino acids, a well-studied chiral system, are used as a panel for identifying structural features and optimal conditions to achieve enantiomer separation with tri-nuclear copper clusters. Several amino acids are also being explored as potential chiral selectors including His, Pro, Arg, Aln, Gln, Tyr, and Trp. Rp-p can be used to quantify the degree of separation and ranged from 0.6 to 1.18 for hydrophobic amino acids in the presence of a histidine chiral selector.
In order to investigate the structural differences between resolvable chiral complexes, a combination of MS/MS, CCS measurements, and molecular mechanics techniques have been investigated. Lead candidate structures are currently being validated with additional experiments and higher-level computational approaches.
Lipid Characterization by Ion Mobility-Mass SpectrometryInvestigator: Katrina Leaptrot, Ph.D. |
 |
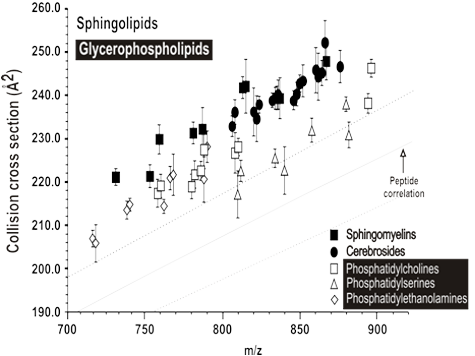
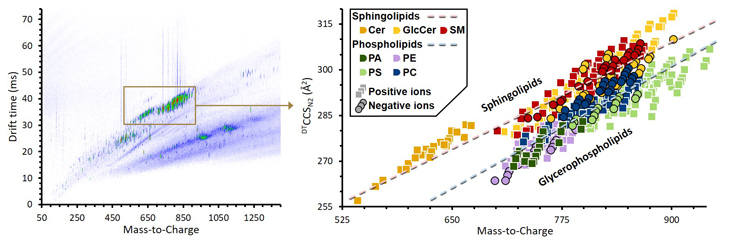
Lipids are an essential class of biomolecules that are involved in a wide range of biological processes and are associated with numerous diseases. These molecules are structurally diverse; varying in headgroup type, acyl chain length, position of attachment, degree of unsaturation, and stereochemistry. Many lipids are isobaric, making them indistinguishable by mass spectrometry alone. Using ion mobility to complement the mass analysis provides information on size, shape (collision cross section, CCS), and mass-to-charge (m/z) that is selective to gas-phase structure. Ion mobility-mass spectrometry (IM-MS) can delineate biomolecules into their respective classes and differentiate isomers.
We compiled a structural database of 456 sphingolipid and glycerophospholipids species and observed predictable changes in CCS for related lipids, with sphingolipids exhibiting 2-6% larger CCSs than glycerophospholipids of similar mass. Of the two primary structural descriptors found to affect CCS, changes in degree of unsaturation were found to be four times as influential as changes in acyl tail length. Relative CCS changes were found to be generally commensurate with adduct size, with larger adducts resulting in larger CCSs. The quantitative trends developed in this work are expected to aid in identifications of lipid features in future lipidomics analyses.
(1) Leaptrot, K. L.; May, J. C.; Dodds, J. N.; McLean, J. A., Nature Communications 2019, 10 (1), 985.
Ion Mobility Resolution of Challenging Isomer Systems using High Resolution Demultiplexing: HRdmInvestigator: Jody C. May, Ph.D. |
 |
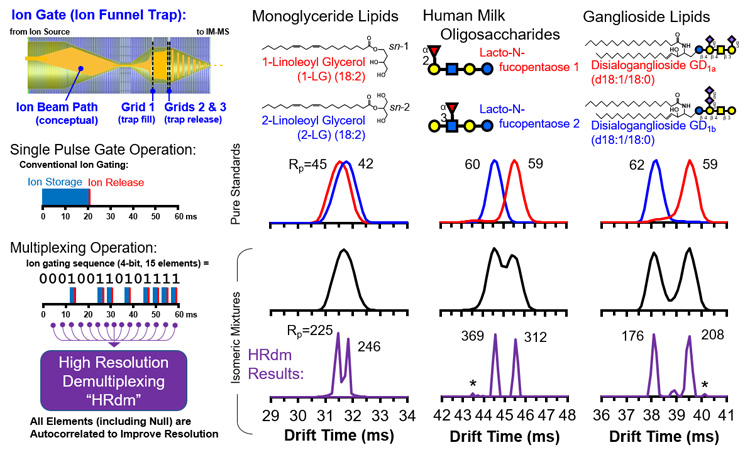
Ion mobility separates ions based upon their effective size in the gas phase, and thus can differentiate isomers which have identical masses and thus cannot be separated by conventional mass spectrometry alone. For challenging isomer systems in which size differences are small, conventional ion mobility is oftentimes not enough to resolve isomeric peaks.
To address this, a new approach based upon a combined ion multiplexing and post-acquisition peak deconvolution strategy has been developed in collaboration with Agilent Technologies. Referred to as HRdm, this approach can achieve ion mobility resolving powers of greater than 200 at the highest processing level and has been demonstrated to resolve challenging isomeric systems. Currently this strategy is being optimized and applied to various isomeric systems in order to establish the application space for this technique.
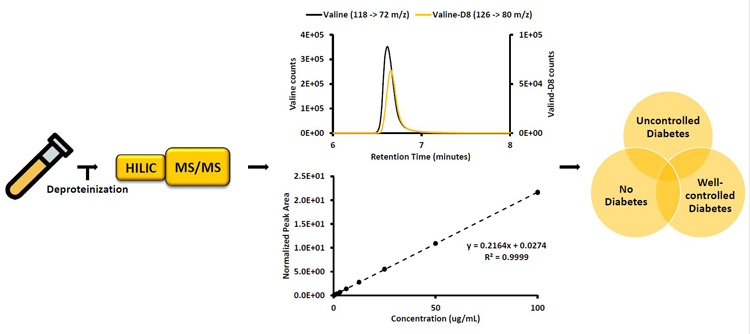
Quantitation of 11 Amino Acids in Human Plasma by LC-MS/MS: Applications in the Prediction of PrediabetesInvestigator: Amelia L. Taylor |
 |
While type II diabetes affects many individuals worldwide and has many negative consequences, current diagnostic tests only detect diabetes after irreversible negative effects may have taken root. Therefore, this work focuses on developing a novel early biomarker of diabetic disease state by quantifying free amino acids circulating in plasma using liquid chromatography-tandem mass spectrometry (LC-MS/MS), as previous results suggest that these molecules are in a state of imbalance in prediabetes and type II diabetes.
Using hydrophilic interaction liquid chromatography (HILIC), a novel method was developed and validated (according to the FDA’s Bioanalytical Method Validation Guidelines) to quantitate 11 amino acids from human plasma (Aln, Glu, Gly, Ile, Leu, Met, Phe, Thr, Trp, Tyr, and Val). Future work will consist of determining if the amino acids being quantified correlate to diabetic disease state by analyzing samples from patients without diabetes, with well-controlled diabetes, and with uncontrolled diabetes.
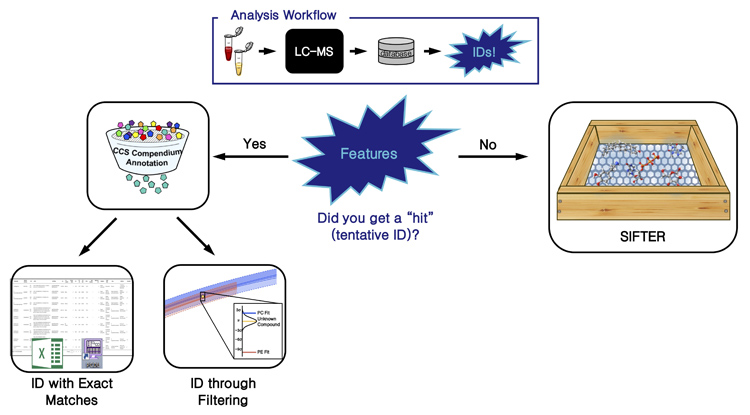
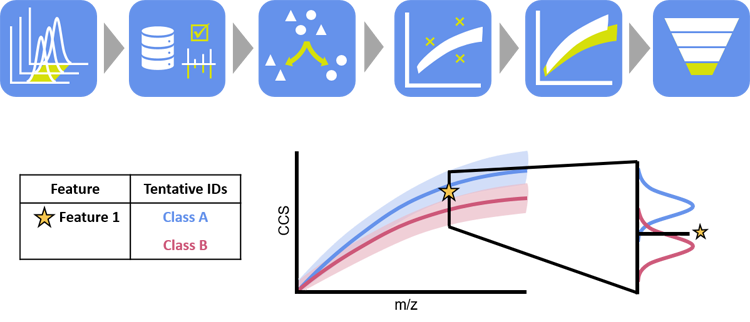
Unified Collision Cross Section Compendium for Compound Annotation and Chemical Class PredictionInvestigator: Jaqueline A. Picache |
 |
To alleviate the burden of manual effort and improve the efficiency of generating accurately annotated, high confidence identifications, my research goals have focused on developing necessary informatics tools to distill the Liquid Chromatography-Ion Mobility-Mass Spectrometry data deluge into operational information and biological inferences. The CCS Compendium is the beginning of this effort and is an open source, collaborative effort and has been designed to be expanded using crowd sourcing techniques.1
Informatic tools have been developed to aid in global, untargeted omic studies. In these studies, the data output is annotated with a commercially and/or in-house database. Some features or compounds can be identified this way, however, most features remain unidentified or have a tentative identification match (multiple candidates based on chemical formula). Using the CCS Compendium, a filtering tool was developed to improve the annotation of these tentative identifications based on class and/or subclass. To address the challenge posed by unidentified features that do not have a database hit, a machine learning algorithm was developed. This algorithm, called SIFTER (Supervised Inference of Feature Taxonomy using Ensemble Randomization), requires m/z, CCS, and Kendrick mass defect as inputs to predict an analyte’s super class, class, and subclass. Briefly, each classification (super class, class, and subclass) is predicted independently and concatenated into a final predictive annotation. Current efforts are focused on incorporating new data into the Compendium and providing additional user-requested informatic tools.
Link to the Unified CCS Compendium
(1) Picache J.A., Rose B.S., Balinski A., Leaptrot K.L., Sherrod S.D., May J.C., McLean J.A. Collision cross section compendium to annotate and predict multi-omic compound identities, Chem. Sci. 2018, 10(4):983-993.
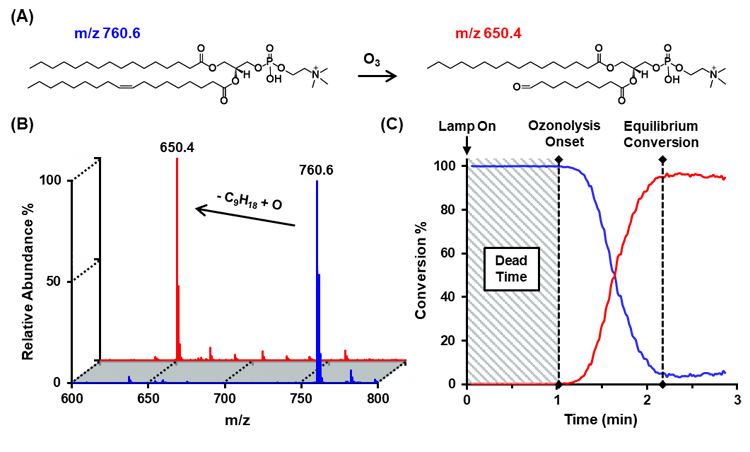
Evaluating New Fragmentation Technologies in Conjunction with Ion Mobility-Mass Spectrometry for Improved Lipid Structural CharacterizationInvestigator: Rachel A. Harris |
 |
Collision induced dissociation (CID) has long served as the workhorse for structural analysis in workflows utilizing mass spectrometry. However, the development of next-generation fragmentation techniques is poised to offer increased specificity and structural information for improved identification of lipids.
Two techniques currently in development include surface induced dissociation (SID) and online ozonolysis of double bonds. Surface induced dissociation, in contrast to collision induced dissociation, imparts a large quantity of energy to analyte molecules in a single, rapid collision step so that dissociation proceeds under kinetic, rather than thermodynamic, control. In this way, SID is able to access fragmentation pathways unavailable to CID and thus yields new structural information. Ozonolysis, on the other hand, involves the reaction of generated ozone in the solution phase with unsaturated species, ultimately leading to cleavage of the double bond. Thus, the position of the double bond in the species can be elucidated, a feat that has proved challenging with other dissociation techniques.
The development of an online ozonolysis system allows for the rapid identification of unsaturated species, and is easily coupled to other mass spectrometric techniques for increased avenues for compound characterization, such as ion mobility.
Biomarker Discovery and Validation for Delirium Syndrome using Mass Spectrometry-based Metabolomics Analysis of Human Serum SamplesInvestigator: Don E. Davis, Jr. |
 |
Delirium is a heterogeneous, neuropsychiatric syndrome characterized by inattention and acute loss of cognition and is subtyped by psychomotor activity based on different heterogeneity factors, which makes it difficult to characterize. Psychomotor activity is based on the patient’s motor activity, speech, and level of arousal. Subtyping is critical to determine appropriate clinical intervention. The two major psychomotor subtypes of delirium are 1) hypoactive and 2) normoactive:
- Hypoactive delirium is described as quiet delirium and patients with this subtype appear drowsy, somnolent or lethargic. Because its clinical presentation can be very subtle, hypoactive delirium is frequently undetected by health care providers and may be ascribed to other causes such as depression or fatigue. At the same time, this type of delirium is considered to portend the worst prognosis and is associated with higher short-term and long-term mortality.
- In normoactive delirium a patient’s responsiveness to the environment is normal, neither decreased or increased arousal. The most commonly studied delirium subtype is based upon the patient’s psychomotor activity.
Categorizing delirium by its psychomotor subtype is complex and is not standardized, which limits its reliability and clinical utility. Nevertheless, most delirium screening tools used currently in research and clinical practice dichotomize its diagnosis into presence or absence, making identification of potentially important subtypes difficult. We hypothesize there is a metabolic difference in psychomotor subtypes of delirium (hypoactive and normoactive).
A Validated, Clinically Quantitative LC-MS/MS Method and a Qualitative LC-DTIM-MS Method for the Analysis of Anti-Epileptic Drugs (AEDs) in Human SerumInvestigator: Don E. Davis, Jr. |
 |
Routine small molecule quantification in the clinical setting is challenging. Some commonly employed methods, such as immunoassays and liquid chromatography with ultra violet (UV) detection, may not exhibit the selectivity or low limits of quantification required for clinical analysis. This project focuses on a reversed phase liquid chromatography-tandem mass spectrometry (RPLC-MS/MS) method to detect and quantify 10 anti-epileptic drugs (AEDs) in human serum. Calibration curves for each AED were generated to assess the limit of quantification (LOQ). The optimized method described herein was validated in accordance with the U.S. Food and Drug Administration (FDA) Bioanalytical Method Validation Guidance for Industry and evaluates AED analyte recovery, matrix effects, precision, accuracy, selectivity, stability, and carry-over. Also, this project demonstrates the utility of liquid chromatography drift tube ion mobility mass spectrometry (LC-DTIM-MS) for increasing confidence in AED identification through the use of accurate mass and drift time/collision cross section (CCS) measurements. Finally, this project provides clinical and research laboratories with a robust and selective, quantitative and qualitative protocol for measuring AEDs in human serum.
When methods are applied in the clinic, the results can provide clinicians with a reproducible quantitative method necessary to multiplex AEDs in a single assay. Multiplexing AED measurements by RPLC-MS/MS and LC-DTIM-MS have the potential to enable better optimization of dosing and response to therapy.
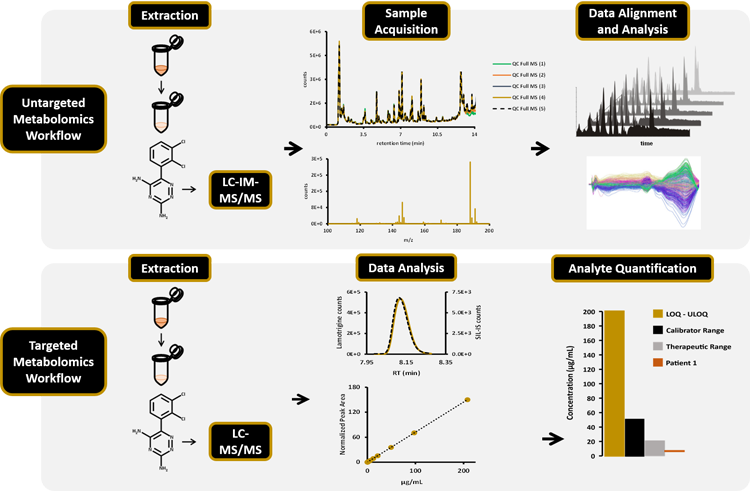
Targeted Workflow: 1) Sample extraction: metabolites extracted by protein precipitation from patient’s serum samples. 2. Sample Acquisition: metabolites analyzed by LC-MS/MS. 3. Data analysis: standard curves generated by extracting normalized peak areas of analyte standards Analyte quantification (targeted): calculate [analyte] in patient samples.
An Integrated LC-IM-MS/MS Workflow for High Confidence Annotations in Untargeted LipidomicsInvestigator: Bailey S. Rose |
 |
Due to their significant role in highly varied cellular functions, a global analysis of lipids is critical for understanding metabolic state. However, the structural diversity and high isomeric prevalence of lipids incurs challenges in the confident annotation of spectral features by MS alone. Therefore, additional complementary techniques such as liquid chromatography (LC) or, more recently, ion mobility (IM) are often necessary to increase the confidence in untargeted lipidomic feature annotation.
To enhance the utility of ion mobility-derived collision cross section (CCS) for characterization, the Unified CCS Compendium has been used in conjunction with the predictive capabilities of statistical programming and regression strategies to quantify class-specific structural trends with the goal of expanding the coverage and confidence of global untargeted studies. To this end, an integrated liquid chromatography-ion mobility-tandem mass spectrometry workflow has been developed to support high-confidence lipidomics. In the workflow, the quantified trends are used to automatically filter annotations to yield higher confidence in the resulting structural assignments. This strategy is being applied to biological problems and systems to enable more complete and straightforward interpretations of the processes implicated in lipid dysregulation.
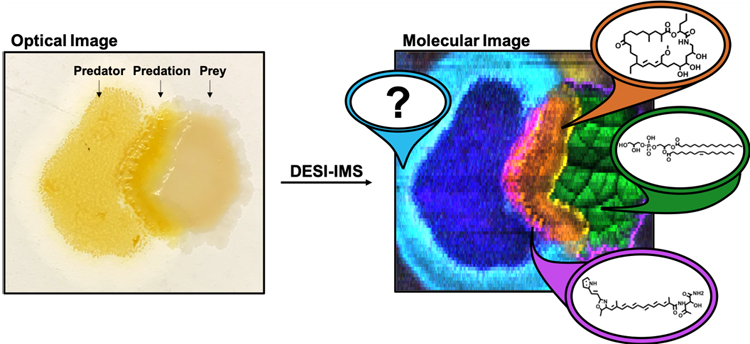
Bacteria Fight Club: Spatiochemically Profiling Intermicrobial RelationsInvestigator: Berkley M. Ellis |
 |
Microorganisms antagonize, manipulate and collaborate with each other and their animal hosts using small molecule metabolites. In human-pathogen interactions, E. coli secreted colibactin intercalates host DNA. In interspecies Staphylocococcus interactions, an anti-MRSA metabolite lugdunin is produced. Natural product secondary metabolites like lugdunin are a prolific source of therapeutic compounds. Therefore, methods that effectively map microbe-microbe interactions will reveal new biology and highlight new therapeutic leads.
To accomplish this feat, we developed an untargeted desorption electrospray ionization- ion mobility imaging mass spectrometry (DESI-IM-IMS) method to spatially annotate excreted metabolites within microbial co-cultures. We apply unsupervised segmentation to acquired IMS data to unbiasedly assess the ecological role of secondary metabolites and spatially prioritize novel natural products.
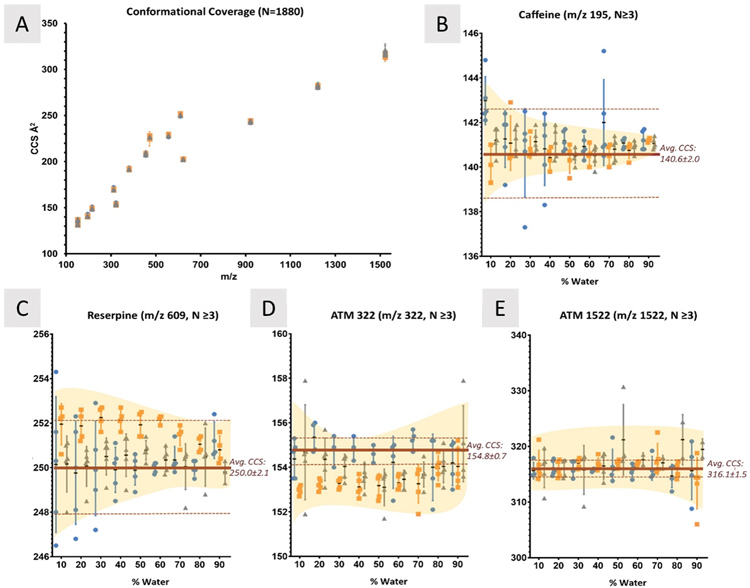
Effects of Solvent and Source Temperature on Ion Mobility Collision Cross Section MeasurementsInvestigator: Nadjali A. Chung |
 |
Ion mobility (IM) is a gas-phase electrophoretic technique that separates ions on the basis of size, shape, and charge, reflected in the IM-derived collision cross section (CCS) value. CCS values represent the rotationally averaged surface area of ions and were found to be reproducible and robust when compared across different low-field drift tube IM (DTIMS)-MS systems.
Recent works have taken advantage of the high reproducibility of CCS values to compile libraries in support of compound identification. To effectively use these CCS libraries, IM parameters need to be standardized, accounting for sources of measurement variation. While many drift tube parameters were assessed in previous work, the effects of solvent and ion source temperatures have not been rigorously documented for small molecules. To address this, this work assessed changes in CCS in different molecular classes across discrete changes in solvent composition and source temperatures.
Selected Past Projects
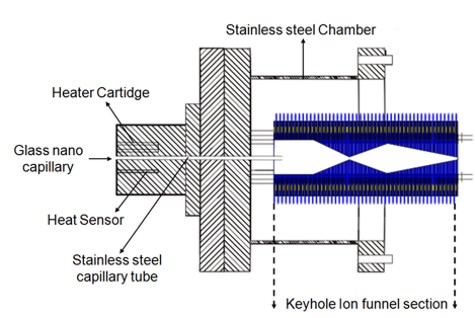
Custom Electrospray Ionization Source
Investigator: Sevugarajan Sundarapandian
Gas-phase protein structure has been extensively studied on many different mass spectrometry platforms. The discrepancy between ESI and MALDI gas-phase structures is an ongoing area of research. For this reason our lab has custom designed and built an electrospray ionization source for our IM-MS instrument. This source features a temperature adjustable stainless steel capillary injection region which permits studies on the temperature dependence of biomolecular structure while also hastening the evaporation of solvent. The keyhole ion funnel section is able to trap and accumulate ions before selectively opening a gating voltage to release the ions into the drift cell, thereby producing a pulsed ion signal.
Characterization of Protein Phosphorylation
Investigator: Randi Gant-Branum
Regulation of protein activity, function, modification, and interaction in the proteome can be attributed in large part to protein phosphorylation. Illnesses associated with altered phosphorylation (i.e.Alzheimer’s disease, cancer proliferation, developmental neurological diseases) can be better understood and treated with more rapid and selective methodologies that identify these symptomatic changes in protein phosphorylation. This work focuses on novel high-throughput methods to site identify, quantitate, and localize phosphorylated peptides and proteins in complex samples such as whole-cell lysates using structural mass spectrometry.
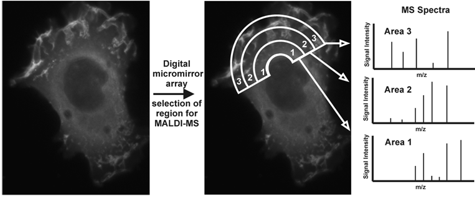
Development of an Optical Device for Laser Shaping and Patterning for Matrix Assisted Laser Desorption/Ionization (MALDI) and its Application in Imaging of Tissues and Cells
Investigator: Michal Kliman
The principal component of the new optical arrangement for MALDI-MS is a digital micro-mirror array (DMA) device. The 1×1.5 cm mirror array of this device consists of ca. one million 13×13 µm size mirrors that can be individually controlled to either reflect or deflect parts of an incoming laser beam. Using this device the MALDI laser can be patterned into regular or complex shapes of variable dimensions and even non-congruent spatial regions can be irradiated simultaneously. Further, the ability to reflect micron sized laser beams allows sub-micron laser focus at the MALDI target without the need for vacuum positioned optics. Prior to reflection from the DMA a laser homogenizer can be used to produce a laser beam with highly uniform photon density to ensure a uniform energy profile across any arbitrary pattern at the target.
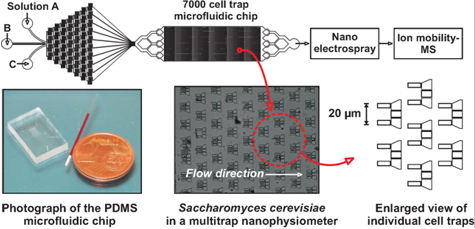
Real Time Analysis of Cellular Effluent to Gauge Cell Signaling and Response
Investigators: Jeff Enders and Cody Goodwin
A critical endeavor in systems biology research is the development of rapid measurement strategies for monitoring comprehensive biomolecular response to changes in chemical environment. To address this challenge, this project utilizes the advantage of combining microfluidic cell trapping with high-throughput real-time biomolecular characterization by nanoelectrospray ionization-ion mobility-mass spectrometry (nESI-IM-MS). In this work, individual cell types are trapped in a microfluidic device and exposed to some form of stimulus (e.g. therapeutic or toxin). Our home built nESI-IM-MS is subsequently used to monitor the dynamics of biomolecular response. Importantly, the use of IM-MS simplifies potentially complicated biomolecular output by performing 2D separations on the basis of both structure and m/z.
Low molecular weight analysis via nanostructure initiator mass spectrometry (NIMS)
Investigators: Jay Forsythe
A shortcoming of traditional matrix-assisted laser desorption ionization (MALDI) mass spectrometry is the tendency for intense matrix background signals to overshadow low mass range analytes.
Analysis of Wound Fluid by Ion Mobility-Mass Spectrometry for Biomolecular Signatures of Diabetic Wound Healing
Investigator: Kelly Hines
Complications of diabetes result in slow or poor healing chronic wounds, which can lead to more serious complications such as infection or amputation. A deeper understanding of the biomolecules involved in the wound healing process may lead to advances in medical treatment for slow healing wounds. Fluid which collects near the site of the wound (i.e., wound fluid) is a non-invasive alternative biological sample for the analysis of biomolecules associated with wound repair. The structural separation afforded by ion mobility-mass spectrometry enables the simultaneous analysis of several classes of biomolecules in a complex biological sample such as wound fluid.
Periodic Focusing DC Ion Guide Development
Investigators: Seth Byers
Recent advances in instrument hardware and software have lead to growth in accessibility and interest in Ion Mobility-Mass Spectrometry experiments over the past two decades. While the MS portion of such devices has been improved to yield staggering mass resolution values, advances in ion mobility resolution have not been as highly developed. Research is currently examining mechanisms for both increasing resolution and ion transmission through ion mobility drift cells. Radial confinement of ion packets in order to better deliver signal through the IM portion of an IM-MS device has been one such mechanism studied.

Lipid Characterization by Ion Mobility-Mass Spectrometry
Investigators: Michal Kliman
Ion mobility – mass spectrometry (IM-MS) has in the last decade established itself as a viable partner of other MS coupled techniques used in biological analysis. This project characterizes lipids not only by their gas phase mobility, (i.e. the migration time through an atmosphere of neutral gas), but also by their collision cross-section (CCS) or apparent surface area. Empirical CCS measurements can be related to the structure of lipids in the gas phase via computational modeling. Computational modeling in tandem with IM experiments can be used to extract important information about predominant molecular folding dynamics and their effect on the gas phase behavior of different classes of lipids. The ability to resolve lipids from other biological signal, resolve various isobaric lipids by type and by the number of double bonds in fatty acid tails while understanding the structural bases for such separations is the forte of this method.