Background
Mapping Your Life and Everything Else
Presented by Prof. John A. McLean at TEDx Vanderbilt University:
Guiding Theory
Our research focuses on the design, construction, and application of advanced technologies for structural mass spectrometry, in particular, for studies in biomolecular separations, biophysics, chemical and synthetic biology. To identify and structurally characterize the inventory of biomolecules arising from complex samples, we perform multidimensional chemical separations centered around condensed-phase chromatography (GC, LC, SFC) combined with ion mobility-mass spectrometry (IM-MS), the latter of which provides separations on the basis of apparent surface area (ion-neutral collision cross section) and mass-to-charge (m/z), respectively. Biomolecular structural information is interpreted by comparing experimentally obtained collision cross-sections in the context of those obtained via predictions from empirical models, machine learning, and molecular dynamics simulations.
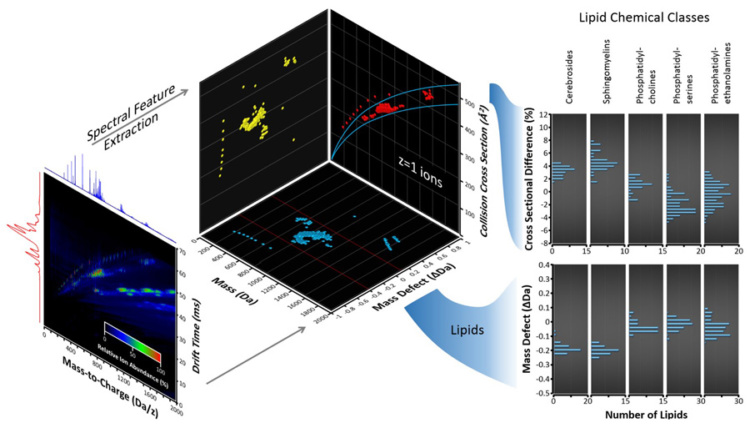
1JC May and JA McLean, Advanced multidimensional separations in mass spectrometry: navigating the big data deluge. Annual Review of Analytical Chemistry 9:387-409 (2016).