Identifying Cell-Specific Transcripts
More is known about the cellular architecture and development of C. elegans than of any other metazoan. For example, the lineal origin and morphology of all 959 somatic cells are fully established. In addition, a wiring diagram traces every synapse among the 302 neurons in the worm nervous system.
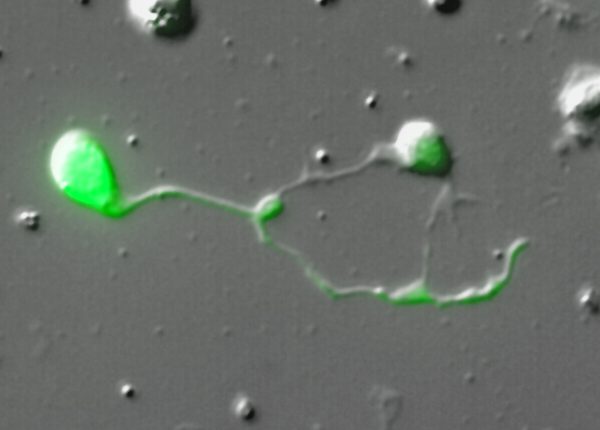 Although the genome is completely sequenced and all active genes have been defined by direct transcript analysis, we lack a gene expression map that matches the single cell resolution of the C. elegans anatomy.
Although the genome is completely sequenced and all active genes have been defined by direct transcript analysis, we lack a gene expression map that matches the single cell resolution of the C. elegans anatomy.
Over a period of several years, the Miller Lab has worked to develop cell-specific profiling methods that can be used to meet this goal. Initially, we helped develop a robust method for culturing C. elegans embryonic cells (Christensen et al., 2001) and then used this approach to isolate specific neurons (Fox et al., 2005) and body muscle cells (Fox et al., 2007) by FACS for microarray profiling studies.
To profile larval and adult cells, we implemented the mRNA tagging strategy to obtain the first comprehensive gene expression profile of the entire C. elegans nervous system (Von Stetina and Watson et al., 2007). Using a combination of these cell-specific RNA isolation methods, the Miller lab joined the modENCODE project to define all C. eleganstranscripts (Gerstein et al., 2010, Gerstein et al., 2014). For this purpose, we isolated RNA from over 30 cell types and larval stages and used whole genome tiling arrays to detect transcripts. A comprehensive analysis of these data sets revealed cohorts of genes that are differentially expressed across cell types and detected shared regulatory motifs with potential roles in gene regulation (Spencer and Zeller et al., 2011)(Hallem et al., 2011).
 A surprisingly large number of differentially expressed non-coding RNAs were also detected (Lu et al., 2011). All of these data are available here along with tools for visualizing transcripts (Genome Browser) and for quantifying expression levels for specific genes across cell types and developmental stages (WormViz).
A surprisingly large number of differentially expressed non-coding RNAs were also detected (Lu et al., 2011). All of these data are available here along with tools for visualizing transcripts (Genome Browser) and for quantifying expression levels for specific genes across cell types and developmental stages (WormViz).
In addition to cataloging gene expression fingerprints for individual cell types, we have also used our profiling methods to identify the targets of transcription factors that regulate regulate key mechanisms in neural development such as synaptic specificity (UNC-4) (Von Stetina and Fox et al., 2007), circuit remodeling (UNC-55, IRX-1) (Petersen et al., 2011) and dendritic branching (MEC-3) (Smith and O’Brien et al., 2013).
Specific neurons are accessible to isolation by FACS from multiple larval stages
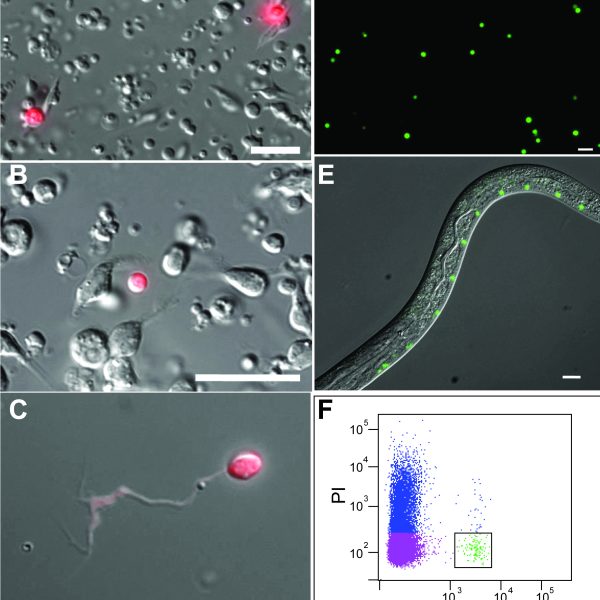 We have recently developed methods for generating cell-specific RNA-Seq profiles from C. elegans larvae and adults (Spencer et al., 2014). In the SeqCel (RNA Seq of C. elegans cells) method, we use FACS to isolate fluorescently labeled cells for RNA-seq analysis.
We have recently developed methods for generating cell-specific RNA-Seq profiles from C. elegans larvae and adults (Spencer et al., 2014). In the SeqCel (RNA Seq of C. elegans cells) method, we use FACS to isolate fluorescently labeled cells for RNA-seq analysis.
In our initial studies, we have focused on neurons because the nervous system contains the largest variety of specific cell types in C. elegans. Applications include neuron-specific profiling (Lim et al, 2016; Oranth et al., 2018) and identifying transcriptionally-regulated genes for neuron regeneration (Byrne et al., 2016).
C. elegans Neuron Gene Expression map and Network (CeNGEN).
CeNGEN is also striving to produce whole transcriptome (“bulk”) RNA-Seq data for all C. elegans neurons. To achieve this goal, we are fluorescent markers that can be used to label each of the 118 known types of neurons for isolation by FACS.
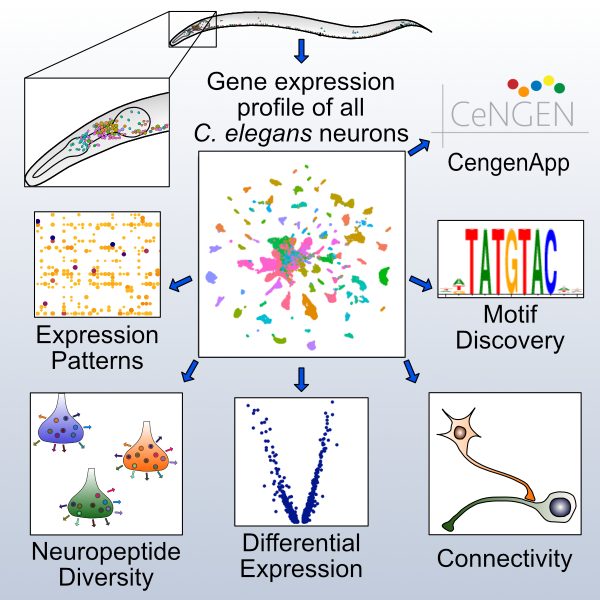
This approach is designed to catalog neuron specific alternative splicing and to capture noncoding RNAs. Additional ongoing projects include the analysis of the male nervous system to identify sex-specific transcripts and to catalogue neuron-specific developmental regulation of gene expression.