Research Focus
Below is a summary of the projects currently within the Iverson lab. To read a more in-depth research summary, click here.
Signaling in mammals
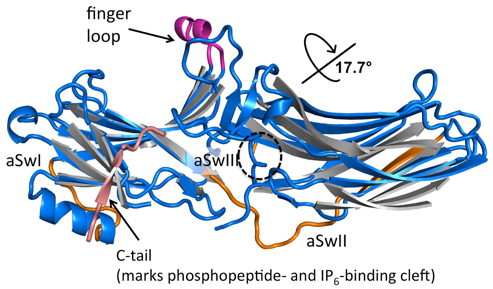
Figure 1: Arrestin activation. Overlay of the N-domain of IP6-activated arrestin-3 (blue) with the secondary structural elements of basal arrestin-3 (grey with a pink C-tail) highlights the interdomain rotation. Activating sites include the finger loop, with the active, alpha-helical conformation in magenta, and phosphate binding to the cleft that binds C-tail in the basal conformation. Activation induces previously undescribed conformational changes in the distal aSw regions (orange). We hypothesize this is required for effector activation.
Our largest focus in the laboratory involves signaling mediators called arrestins (GM120569, DA043680). Arrestins were first discovered for their ability to bind active, phosphorylated GPCRs and suppress G protein-mediated signaling. Subsequent findings suggested that the receptor-bound form was in an activated state and initiates a second, G protein-independent wave of signaling. The conformations of free (basal) and active arrestins are quite different (Fig. 1). Nevertheless, major questions remain on how activated arrestins promote signaling.
Complex II
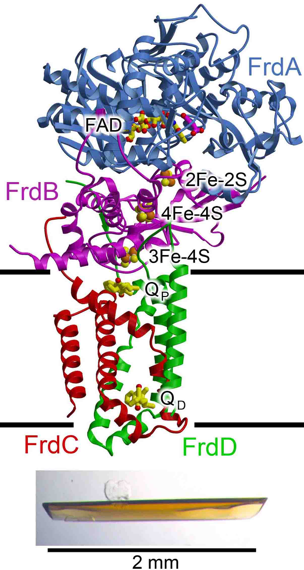
Figure 2: E. coli QFR. The complex contains four polypeptides (FrdABCD). In the figure, the flavoprotein is cyan, the iron protein is red, and the membrane proteins are colored green location of the membrane.
Complex II catalyzes the oxidation of succinate to fumarate during the citric acid cycle and passes the two electrons from this reaction to membrane-soluble quinones. Members of the complex II family contain a soluble region consisting of two polypeptide chains (flavoprotein and iron protein), and a poorly-conserved membrane-spanning domain. Quinol-fumarate reductase (QFR) is a complex II homolog that catalyzes the reduction of fumarate to succinate during anaerobic respiration.
We are currently performing several exploratory avenues of research (GM061606). The first is how this complicated respiratory protein is assembled, including how the cofactors are inserted. Recent studies have focused on the insertion of the flavin cofactor and the role of newly discovered assembly factors in this process. The second avenue of research is how the E. coli QFR affects chemotaxis (Fig. 2; [34]). Both QFR and its metabolite fumarate are required for cellular locomotion, and QFR controls the onset of tumbling. Our final avenue in this research is focused on the human enzyme, with the long-term goal of understanding how disease-associated mutations affect the biochemical properties of the enzyme. The research on the human enzyme is in the early stages, and we remain focused on methods to express the properly assembled complex.
Pathogen-host molecular recognition
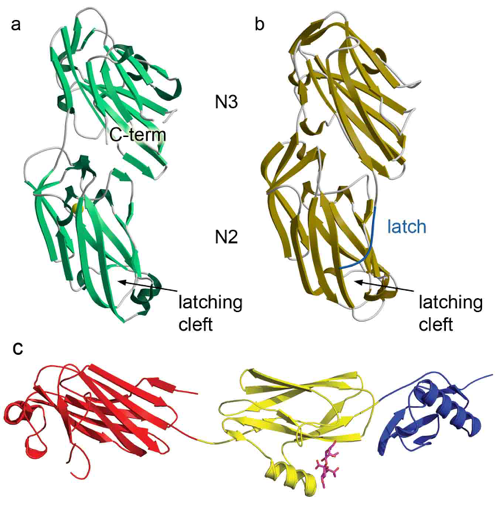
Figure 3: Structures of SRR adhesins. (a) Srr1 from S. agalactiae. (b) Srr2 from S. agalactiae. (c) GspB from S. gordonii colored by domain.
Our present investigations focus on the interaction between human platelets and the Serine Rich Repeat (SRR) adhesin family (AI106987). These proteins are required for the development of infective endocarditis, which is a life threatening infection associated with an in-hospital mortality rate of ~20%, and a 5-year mortality rate of ~40-70%, depending on the causative organism. Indeed, despite prompt therapy, patients of streptococcal endocarditis have a poor prognosis: 50-60% of patients undergo heart failure or progressive valve destruction, and ~15% of patients undergo systemic embolization and stroke. Treatment for bacterial endocarditis commonly combines an antibiotic regimen with surgical intervention, with both having shortcomings. Our research investigates the basic mechanisms of bacterial attachment to the host.