Mapping Antibody-Antigen Interactions
The human immune system creates antibodies in response to viral threats such as SARS Coronavirus 2 (SARS-CoV-2), influenza (Flu), human immunodeficiency virus (HIV), or Ebola/Marburg viruses. These antibodies recognize proteins on the surface of the virus, so-called antigens. In the best-case scenario, human antibodies neutralize viruses by blocking the interaction of viruses with the surface of the human cell. Viral vaccines mimic the effect of infection to provoke our immune system to elicit neutralizing antibodies, thereby preventing infection during later exposures. Unfortunately, not all humans create neutralizing antibodies quickly enough upon infection, and there are no effective vaccines available for many pathogens.
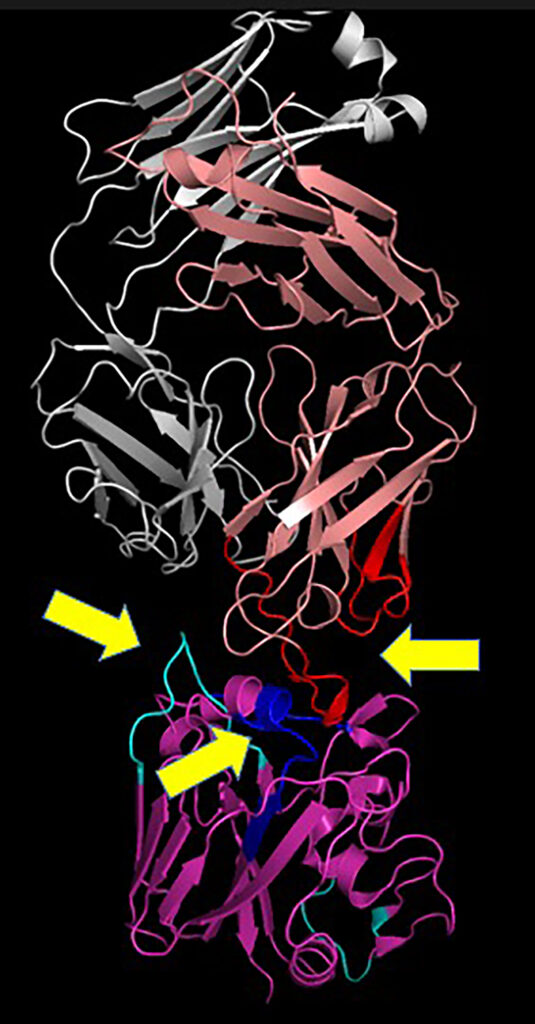
The Schey lab is collaborating with the laboratories of Dr. James Crowe III and Dr. Jens Meiler at Vanderbilt to use mass spectrometry tools to determine antibody-antigen interactions. Tools such as hydrogen-deuterium exchange mass spectrometry (HDX-MS) and crosslinking mass spectrometry (XL-MS) are being used to examine a variety of antibodies to proteins from viruses mentioned above. Knowledge of epitope-paratope interactions will inform on developing new therapeutic antibodies for treating viral infections.
Relevant References
Tran MH, Schoeder CT, Schey KL, Meiler J. Computational structure prediction for antibody-antigen complexes from hydrogen-deuterium exchange mass spectrometry: Challenges and Outlook. Front. Immunol., 13:859964, 2022.
Doyle MP, Kose N, Borisevich V, Binshtein E, Amaya M, Nagel M, Annand EJ, Armstrong E, Bombardi R, Dong J, Setliff I, Georgiev I, Schey KL, Broder CC, Geisbert TW, Cross RW, Crowe Jr, JE. Functional cooperativity mediated by rationally selected combinations of human monoclonal antibodies targeting the henipavirus receptor binding protein. Cell Reports, 36:109628, 2021.
Williamson L, Reeder K, Bailey K, Tran M, Roy V, Fouch M, Kose N, Trivette A, Nargi R, Winkler E, Kim A, Gainza C, Rodriguez J, Armstrong E, Sutton R, Reidy J, Carnahan R, McDonald WH, Schoeder CT, Klimstra W, Davidson E, Doranze B, Alter G, Meiler J, Schey KL, Julander J, Diamond M, Crowe JE. Therapeutic alphavirus cross-reactive E1 human antibodies inhibit viral egress, Cell, 184:4430-4446, 2021.
Morris AK, Wang Z, Ivey AL, Xie Y, Hill PS, Schey KL, Ren Y. Cellular mRNA export factor UAP56 recognizes nucleic acid binding site of influenza virus NP protein. Biochem. Biophys. Res. Commun., 525:259-264, 2020.
Bennett MR, Dong J, Bombardi RG, Soto C, Parrington HM, Nargi RS, Schoeder CT, Nagel MB, Schey KL, Meiler J, Skaar EP, Crowe Jr JE. Human VH1-69 Gene-encoded human mAbs against Staphylococcus aureus IsdB use at least three distinct modes of binding to inhibit bacterial growth and pathogenesis. mBio, 10:e02473-19, 2019.
Bennett MR, Bombardi RG, Kose N, Parrish EH, Nagel MB, Petit III RA, Read TD, ScheyKL, Thomsen IP, Skaar EP, Crowe Jr JE. Human mAbs to Staphylococcus aureus IsdA Provide Protection Through Both Heme-Blocking and Fc-Medicated Mechanisms. J. Infect. Dis., 219:1264-1273, 2019.
Sevy AM, Wu NC, Gilchuk IM, Parrish EH, Burger S, Yousif D, Nagel MBM, Schey KL, Wilson IA, Crowe Jr JE, Meiler J. Multistate design of influenza antibodies improves affinity and breadth against seasonal viruses. Proc. Natl. Acad. Sci., 116:1597-1602, 2019.