Research
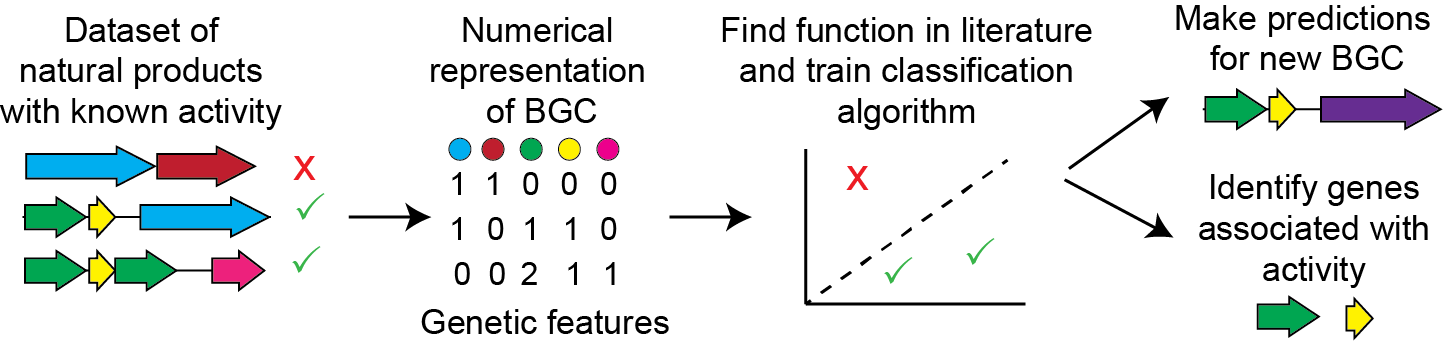
Predicting natural product activity from biosynthetic gene cluster sequences
Natural products are an excellent source of bioactive molecules that can be used as therapeutics. Natural product chemists have become increasingly adept at using genomic data to infer natural product structure and to discover novel molecules. However, these molecules are not always active and there is currently no general and reliable method to predict function from a biosynthetic gene cluster (BGC). We recently developed a machine learning algorithm predicting natural product antimicrobial or antitumor activity based on the sequence of the biosynthetic gene clusters (BGCs). Bioinformatics analysis is fast and inexpensive whereas production, purification, activity assay, and structural elucidation are time consuming, creating bottlenecks in natural product discovery. Our algorithm should make it possible to prioritize BGCs that are likely to produce natural products with desirable activities, reducing the number of times experiments need to be passed to discover hits. Currently, we are focused on improving the algorithm and experimentally validating its predictions. We are also working on developing bioinformatics methods to predict small molecule inducers of secondary metabolism in order to accelerate the rate of discovery of natural products from genome mining.
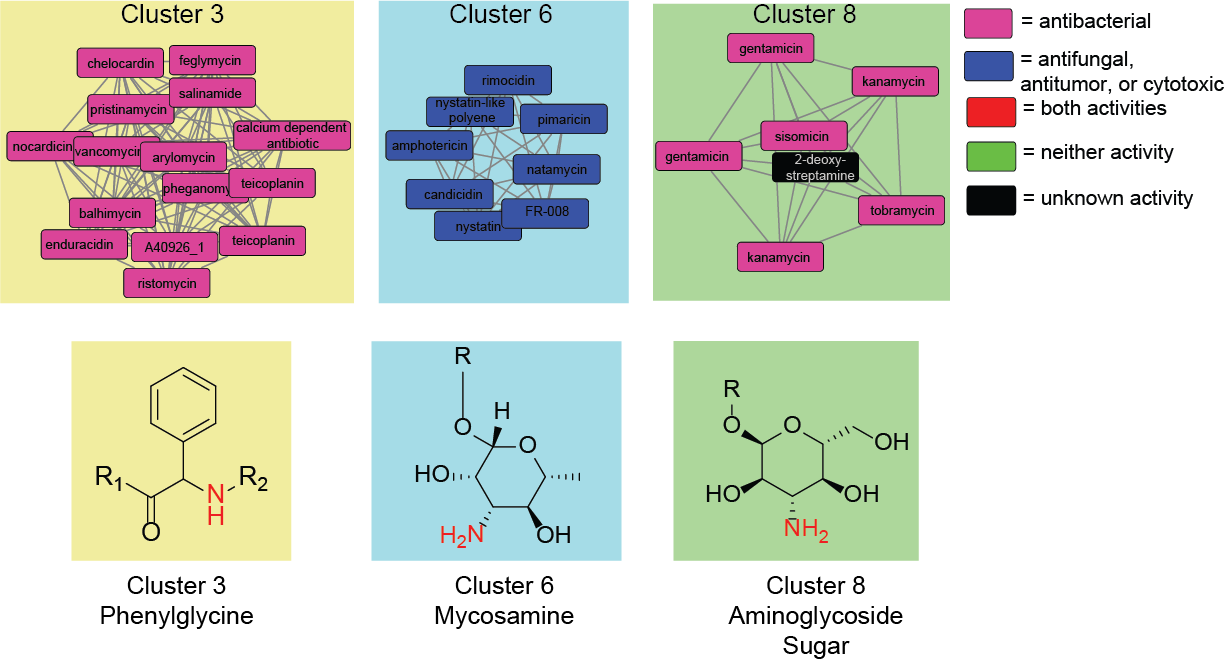
Genome mining for natural products with activity-associated biosynthetic genes
During the development of our machine learning method for predicting natural product bioactivities, we discovered several biosynthetic genes that were associated with activity. By studying novel biosynthetic gene clusters containing activity-associated genes we are investigating whether these biosynthetic genes are useful as genome mining markers. If a gene is validated as being associated with activity, we will then further investigate how the molecular substructure installed by the gene contributes to the natural product’s activity.
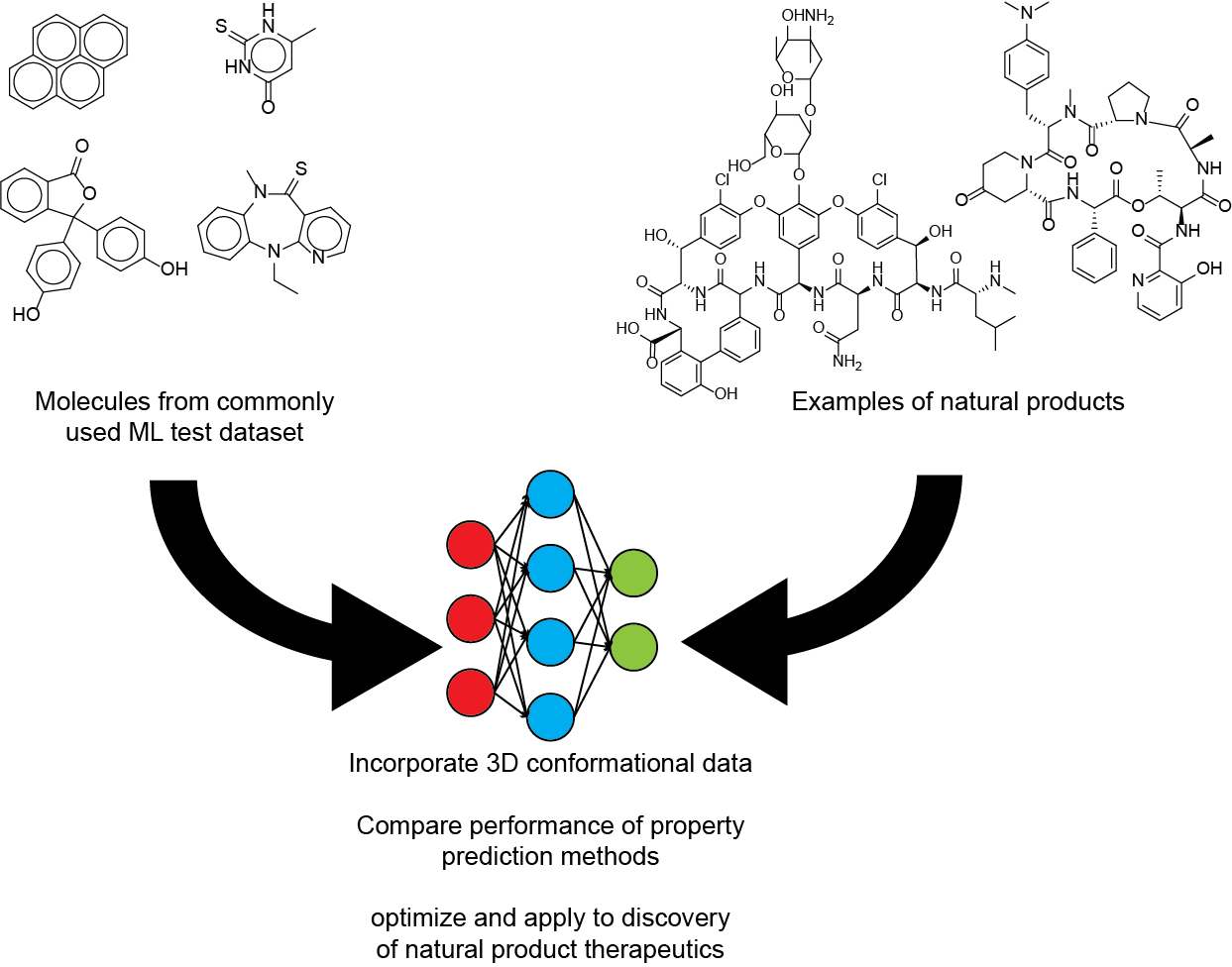
Prediction of molecular properties for natural products and other complex organic molecules
Machine learning algorithms have been very successful at predicting various molecular properties, including those important for drug development such as pharmacokinetic properties, toxicity, and binding to protein targets. Many datasets used for training models that predict molecular properties are focused on relatively small organic molecules that are not as large or complex as common molecules in many natural product classes, specifically non-ribosomal peptides, ribosomal peptides, and polyketides. We are evaluating the accuracy of various machine learning models on natural products and optimizing them to work specifically for natural products or other complex organic molecules. Ultimately, we hope to use these algorithms to assess whether newly discovered molecules may be useful as therapeutics and what chemical modifications would improve their pharmacokinetic properties.
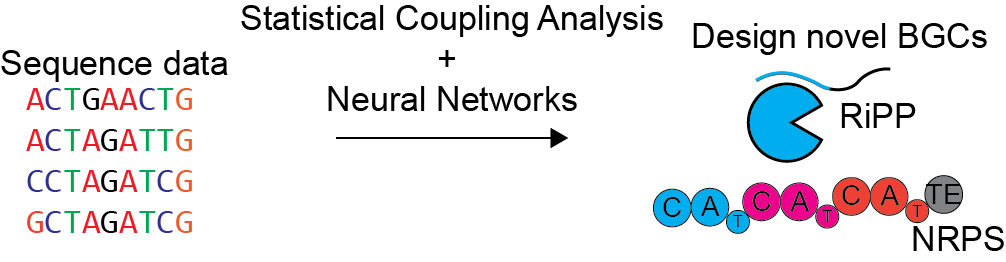
Machine learning-guided synthetic biology
Naturally occurring molecular scaffolds, such as peptides and polyketides, often have desirable bioactivities, but can be difficult to synthesize. The ability to repurpose natural biosynthetic machinery would make these molecules more accessible. Despite several successful attempts at reengineering specific BGCs, there are currently no general strategies for designing biosynthetic pathways to make a specific molecule of interest. We are developing machine learning and statistical models to elucidate rules for engineering BGCs, particularly RiPPs, NRPS, and PKS pathways and then apply these rules to produce novel bioactive molecules. We will experimentally validate our machine learning models using heterologous expression.
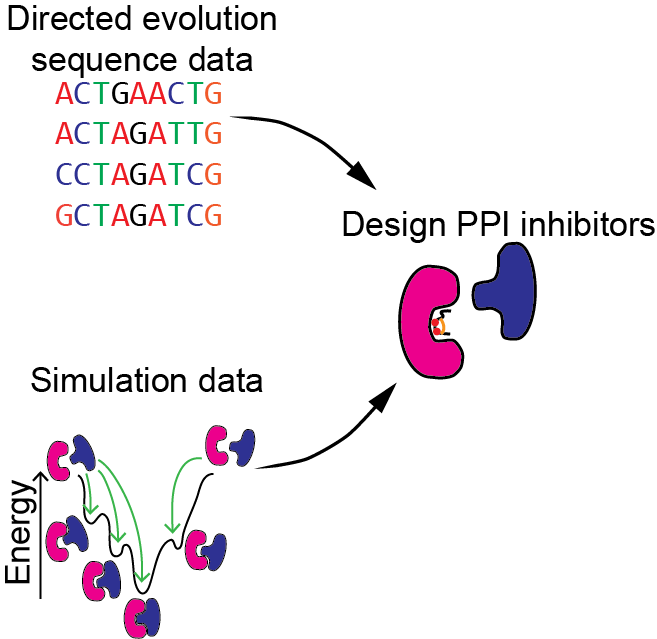
Machine learning for the design of peptide-based protein-protein interaction inhibitors
Protein-protein interactions (PPIs) were long considered “undruggable” but recently proteins, peptides, and peptidomimetics have been used to inhibit PPIs. PPI inhibitors make it possible to target proteins without ligand or substrate binding sites. We will develop machine learning algorithms for designing PPI inhibitors and use directed evolution experiments to collect additional data to improve our machine learning models. We will focus on using ribosomally encoded and post-translationally modified peptides (RiPPs) as PPI inhibitors.