The Hamm Lab
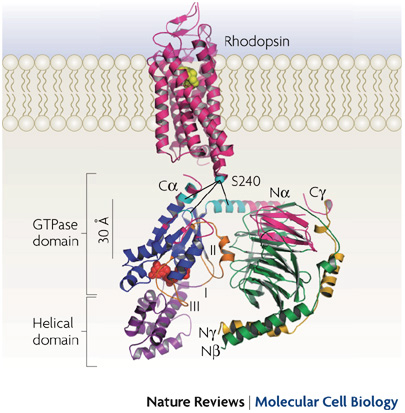 Ribbon model of rhodopsin (Protein Data Bank (PDB) ID 1GZM12) juxtaposed with the Gt heterotrimer (PDB ID 1GOT32). G-protein-coupled receptors (such as rhodopsin; pink) share a common structure with seven-transmembrane-spanning α-helices. Receptor activation exposes the G protein binding site that is formed by the intracellular loops of the receptor. Biochemical studies have identified several receptor contact sites on the G protein (pink and cyan). A few studies have also defined specific point-to-point interactions, such as between Ser240 on intracellular loop 3 of the receptor (cyan sphere) with specific regions of the Gα N terminus (Na), C terminus (Ca) and α4–β6 loop (cyan)66, 70. These contact regions, with the fatty acid modifications on the Gα N terminus and Gγ C terminus, suggest a probable orientation with respect to the membrane. This orientation places GDP (red spheres) in the nucleotide-binding pocket on Gα (blue) ~30 Å from the sites of nearest receptor contact, posing the question of how receptors cause GDP release from this distance.
Ribbon model of rhodopsin (Protein Data Bank (PDB) ID 1GZM12) juxtaposed with the Gt heterotrimer (PDB ID 1GOT32). G-protein-coupled receptors (such as rhodopsin; pink) share a common structure with seven-transmembrane-spanning α-helices. Receptor activation exposes the G protein binding site that is formed by the intracellular loops of the receptor. Biochemical studies have identified several receptor contact sites on the G protein (pink and cyan). A few studies have also defined specific point-to-point interactions, such as between Ser240 on intracellular loop 3 of the receptor (cyan sphere) with specific regions of the Gα N terminus (Na), C terminus (Ca) and α4–β6 loop (cyan)66, 70. These contact regions, with the fatty acid modifications on the Gα N terminus and Gγ C terminus, suggest a probable orientation with respect to the membrane. This orientation places GDP (red spheres) in the nucleotide-binding pocket on Gα (blue) ~30 Å from the sites of nearest receptor contact, posing the question of how receptors cause GDP release from this distance.
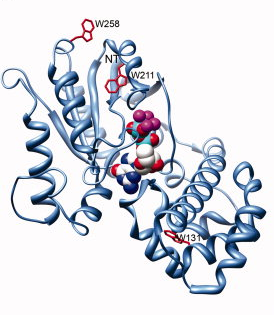
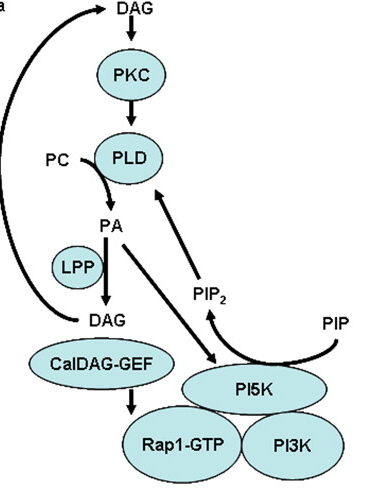
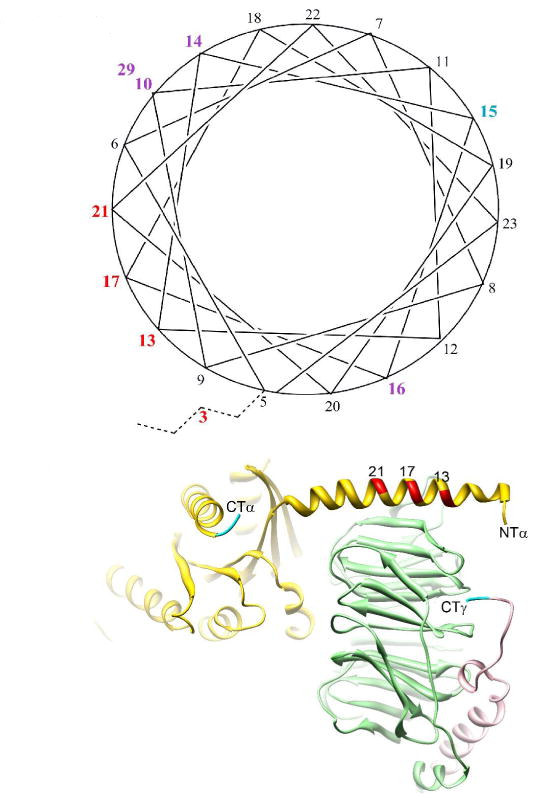
Receptor-mediated changes at the myristoylated amino terminus of Galpha(il) proteins.
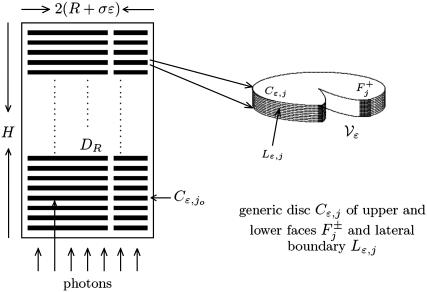
Modeling the role of incisures in vertebrate phototransduction.