OPEN POSITIONS FOR POSTDOCS OR SENIOR SCIENTISTS
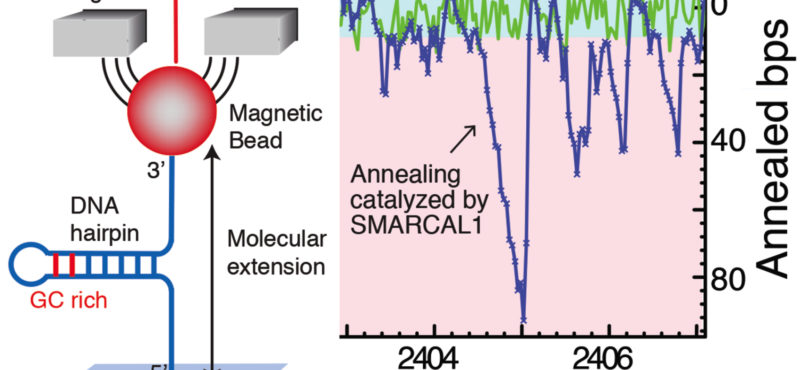
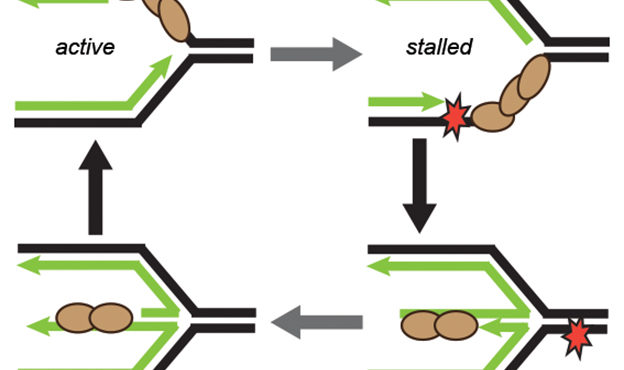
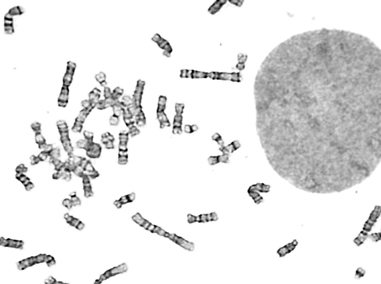
Billions of base pairs of DNA must be replicated trillions of times during a human lifetime. Adding to the difficulty, thousands of DNA lesions happen in each cell of our body every day. Furthermore, replication is also challenged by difficult to replicate sequences and conflicts with transcription. DNA damage and replication stress response mechanisms act to repair the damaged DNA, signal cell cycle checkpoint activation, ensure completion of DNA replication, and maintain genome stability. Defects in these mechanisms cause developmental abnormalities, premature aging, and cancer. Replication errors are the single largest cause of cancer.
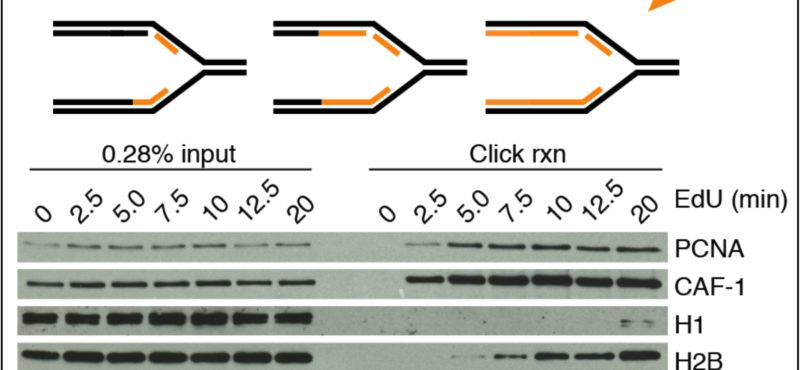
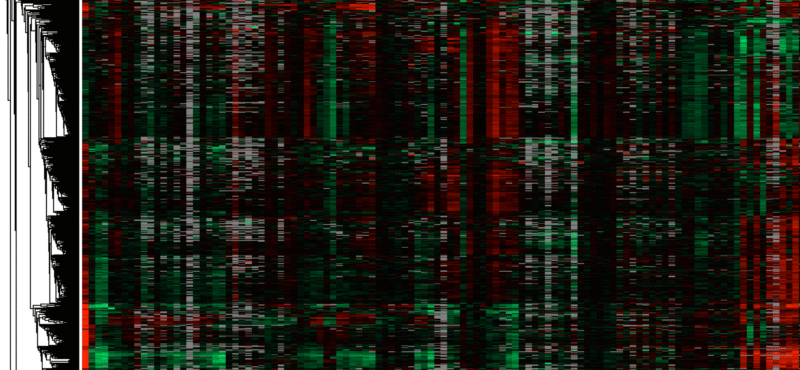
The Cortez lab is dedicated to understanding the mechanisms that maintain genome integrity and promote the complete and accurate replication of the genome. Current projects in the lab include:
- Identification and characterization of new replication stress response proteins
- Mechanistic studies of replication-coupled DNA repair and damage tolerance
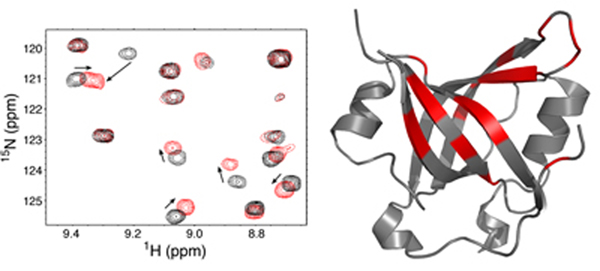
- Analysis of DNA damage signaling pathways controlled by the ATR kinase
- Characterization of abasic site repair and tolerance
- Development of cancer therapeutic approaches targeting the DNA damage response
We use a multi-disciplinary approach including genetics, biochemistry, cell biology, proteomics, and structural biology. The lab is part of the Genome Maintenance Program within the Vanderbilt-Ingram Cancer Center, Vanderbilt Institute of Chemical Biology, and Department of Biochemistry at Vanderbilt University Medical School in Nashville, TN.