Project 1: Modeling the SCLC Phenotype Space
Leaders: Vito Quaranta, Carlos F. Lopez
Co IS: Christine M. Lovly, Jonathan M. Lehman
The overarching goal of this project is to develop a global blueprint of SCLC phenotypic space, clarifying bias imposed by genomic alterations, and epigenetic basis for phenotype transition or selection in response to drugs. We leverage our expertise in modeling the origin of heterogeneous phenotypes from transcription factor (TF) networks inferred from transcriptomics data. Simulations of TF network dynamics via logic-based models enable the identification of attractors, which roughly correspond to SCLC differentiation states defined by profiles of activated or silenced TFs. We define a gene ontology metric to identify biological similarities and differences between phenotypes across a variety of experimental systems including human cell lines, patient-derived xenografts and primary tumors, as well as tumors from SCLC genetically engineered mouse models (GEMMs). The resulting phenotype map informs studies aimed at connecting model systems to patients.
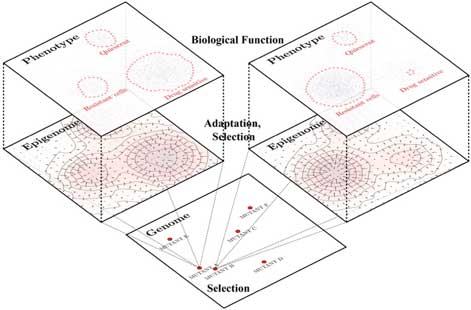
A limitation of SCLC is the scarcity of patient specimens, since biopsies or surgery are rarely performed beyond initial diagnosis. We circumvent this barrier by using instead liquid biopsies of circulating, cell-free DNA as a clinical proxy for the primary tumor, allowing a connection between genomic alterations and phenotypic space states of SCLC tumors. To evaluate the relative role of state transitions vs. selection underneath SCLC plasticity and drug treatment evasion, we use DNA barcoding and information theory techniques to quantify rates of diversification of SCLC phenotypes in response perturbations. Specifically, we map trajectories of cells within the SCLC phenotype space as cells adapt and evade treatment. In summary, we propose to develop a comprehensive view of SCLC phenotypic heterogeneity, linking transcriptomic, genomic, and functional features of SCLC cells across diverse experimental model systems and patient primary tumor specimens. We will link these observations to clinically measurable variables, develop a unified map of phenotypic response dynamics in response to therapy, and analyze the role of plasticity in phenotype shifting and evolution of drug resistance., providing possible novel avenues to SCLC treatment strategies.
