ExRNA
The RNA world and analysis of modern roles for RNA have produced a number of surprises, notably catalytic RNA, the discovery of introns, the prevalence of alternative splicing to generate protein diversity, RNA interference, and riboswitches that link metabolism with gene expression. Recent discoveries showing the extent of transcription within eukaryotic genomes have demonstrated that many RNA species exist in cells, but the exact function of most of these transcripts remains unknown. Many of these RNAs are non-coding. The best-characterized non-coding RNAs are small microRNAs (miRNAs), but many other non-coding RNAs have been identified that appear to play roles in genome defense or chromatin organization. An underlying assumption for many years has been that RNA function is cell autonomous, especially given the levels of RNAse in extracellular fluids and plasma that function to destroy foreign RNAs. However, highly sensitive tools have enabled the discovery of extracellular host RNA in both the bloodstream and multiple body fluids. How these RNAs are exported from donor cells, how they are taken up by and/or targeted to specific recipient cells, and how they are released to function inside recipient cells remain mostly unanswered questions.
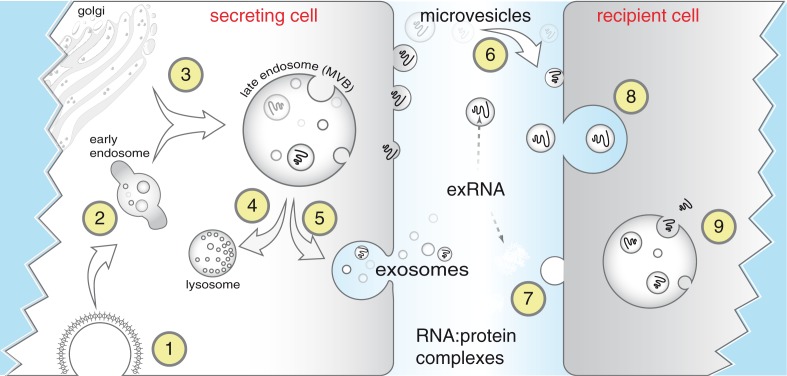
(Patton et al. doi: 10.3402/jev.v4.27494)
Biogenesis and uptake of exRNA. Endocytosis is commonly mediated via clathrin-coated pits (1) after which endocytic vesicles progress from early (2) to late endosomes, also referred to as multivesicular bodies (MVBs). MVBs often fuse with lysosomes for degradation (4) but can also fuse with the plasma membrane (5) thereby releasing exosomes (40–100 nm) to the extracellular space. Microvesicles are larger fragments of plasma membrane that are shed from almost all cells (6). Extracellular RNA (exRNA) can be detected in extracellular vesicles released from secreting cells and can be taken up by recipient cells through receptor-mediated endocytosis (8), by fusion of membranes, or by uptake of RNA–protein complexes or lipoproteins. Although the exact mechanisms remain unclear, exRNA is released inside recipient cells (9) to effect changes in gene expression.