Research
The Meydan Lab is interested in the mechanisms of protein synthesis, and the role of translation regulation and ribosomes in human health. We use various biochemistry techniques, sequencing of ribosome protected mRNA fragments, (a.k.a ribosome profiling (Ribo-seq), disome profiling (Disome-seq), and mitoribosome profiling), and RNA-seq in model systems of Saccharomyces cerevisiae (budding yeast), immortalized human cells, and neurons differentiated from induced pluripotent stem cells (iPSCs) to address our questions.
Research keywords: ribosome, translation regulation, mitoribosome, cellular surveillance, neuronal translation
Current projects in the lab focus on following questions:
1- How are ribosome collisions regulated ?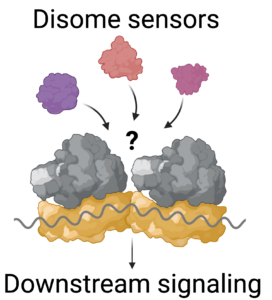
Multiple surveillance mechanisms sense collided ribosomes (disomes) in the cell and lead to distinct cellular outcomes. It is unclear how disome detection and downstream responses are coordinated by different surveillance pathways. The goal of this project is to identify the substrates of different surveillance mechanisms, to establish new methods to detect ribosome collisions in the cell, and to understand how collision levels are regulated.

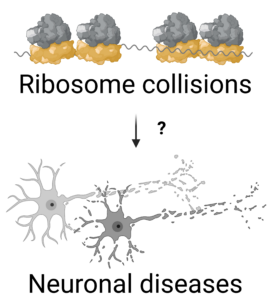
2- How is translation regulated in neurons?
Dysregulation of disome surveillance pathways results in neuronal disease phenotypes. The contribution of disome regulation to neuronal growth and function, however, is not well understood. The goal of this project is to understand how ribosome collisions are regulated in neurons and how disome surveillance pathways contribute to neuronal health.
3- How do disease mutations affect translation elongation?
Many neurodegenerative and neurodevelopmental disease mutations can hinder translation elongation. However, it is unknown if these disease mutations cause ribosome collisions and activate surveillance pathways. The goal of this project is to study the effect of disease mutations on translation elongation and downstream cellular signaling.
4- What are the mechanisms of mitochondrial translation inhibition?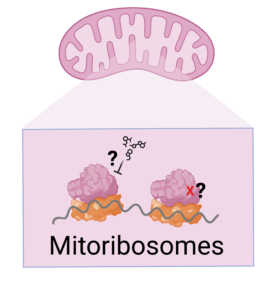
Mitochondria have their own ribosomes (mitoribosomes) for translating mitochondrial proteins. Mitochondrial translation can be hindered by mitochondrial disease mutations and by the off-target activity of antibiotics that are used for inhibiting bacterial translation. The mechanisms of mitochondrial translation inhibition, and overall cellular response to it are unknown. The goal of this project is to identify the consequences of mitochondrial translation inhibition by mitochondrial disease mutations and antibiotics.