Research
Cell Signaling and Immunology

The adaptive immune system is responsible for recognizing foreign antigens and eliciting a response to clear these from our bodies. One important cell type in this response is lymphocytes, which are further divided into B and T lymphocytes. Our lab is particularly interested in T lymphocytes, which house the T-cell receptor (TCR). TCRs specifically recognize cognate 8-10 amino acid long peptides presented among a sea of irrelevant peptides at the surface of antigen-presenting cells. However, the binding affinities of the TCR for these antigens is relatively low, which is unexpected for such a specific interaction. Using single molecule optical tweezers, our lab has demonstrated that external force “energizes” this bond, increasing the lifetime with cognate antigens. Thus, the TCR is a mechanosensor, requiring force to enhance its interaction with specific peptides. Additionally, we showed that applied force decreases the activation threshold for single T-cells by monitoring calcium flux. We are now expanding these investigations into different TCR clonotypes to understand how the mechanical properties of TCRs influence cellular differentiation pathways and subsequent activation.
Cellulose Mechanisms

Cellulose is the world’s most abundant biopolymer known for its high chemical and mechanical stability. It plays a key role in human disease as a component of bacterial biofilms, and is also a source for biofuels. Therefore, understanding how to make and break cellulose impacts diverse aspects of life. Our lab is interested in studying the enzymes that process cellulose, particularly the bacterial cellulose synthase AB complex (BcsAB) and cellobiohydrolase TrCel7A enzymes that make and break cellulose, respectively. Recently, we characterized the activity of single BcsAB molecules under force and over a range of temperatures. Furthermore, we observed that the cellulose strands formed by BcsAB folded back on itself, creating microstructures that impacted polymer mechanics and BcsAB activity. Our work on TrCel7A also suggests that the hydrogen bond network of cellulose influences enzyme activity. Overall, our results indicate that the complex network of interactions surrounding cellulose influence how the productive and destructive enzymatic activities. Better understanding such activities will impact diverse areas, from the treatment/prevention of bacterial infection to more sustainable biofuel production.
Force-Fluorescence Instrumentation
Optical tweezers allow for the precise manipulation of single molecules with nanometer distance and piconewton force resolution. This allows us to probe the molecular reaction coordinate and assess the effects of force on this coordinate. On the other hand, single molecule fluorescence can provide insights into structural transitions in solution. Therefore, combining these two techniques grants unique insight into how molecular machinery harness structural transitions to perform work.
When combining these techniques, one significant challenge has been the high intensity optical trapping laser accelerating photobleaching rates of fluorescent dyes. The laser-induced photobleaching was especially bad for common FRET dyes such as Cy3 and Alexa555. However, we developed an approach to combat this photobleaching, extending fluorescence emission times to over a minute, called Interlaced Optical Force-Fluorescence (IOFF) spectroscopy. Here we provide an example showing the measurement of single molecule DNA unzipping with applied force, coordinated with changes in FRET signals.
Force Generation Mechanism of Kinesin
The Lang lab has published important single molecule data describing the chemomechancial cycle of Kinsein-1, one of the smallest processive biological motors. Our work over the years on Kinesin has been in close collaboration with Dr. Wonmuk Hwang, Dr. Martin Karplus, and Dr. Kristen Verhey. Of note, our data support a mechanism of force generation driven by an unfolded to folded transition between the cover strand and neck-linker of Kinesin-1, forming the cover neck bundle (See Hwang et al. (year), Structure). Furthermore, we directly tested this mechanism by generating kinesin-1 mutants with defects in the regions forming the cover neck bundle. Our results demonstrate cover neck bundle formation is necessary for force generation. The importance of this mechanism was published in PNAS (reference).
Actin Machinery
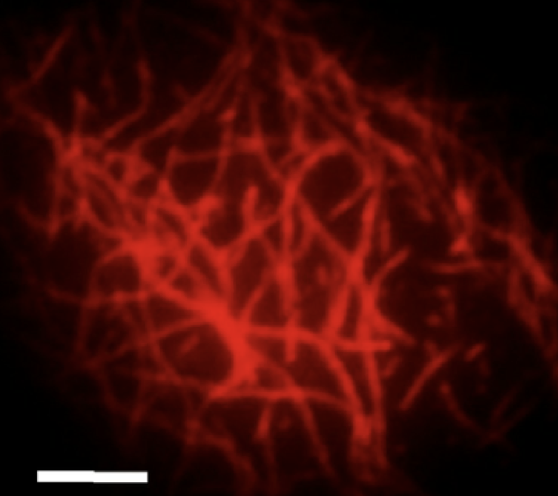
External mechanical forces and forces generated within the cell influence cytoskeletal organization and play a critical role in cell migration, division, growth and apoptosis. Thus, it is important to understand how the structural organization of the cytoskeleton affects the mechanical properties of the cell. In eukaryotic cells, the most abundant cytoskeletal protein is actin. Under specific conditions, monomeric actin polymerizes to form filaments with an approximate diameter of 7-nm and lengths ranging from hundreds of nanometers to few micrometers. In vivo, the organization of F-actin into higher-order structures is regulated by a wide variety of actin binding proteins (ABPs), which are spatially diverse throughout the cell body. For instance, cross-linking proteins localized in the vicinity of the plasma membrane, like filamin, promote the formation of an isotropic F-actin network known as the cortex, which provides and maintains the structural integrity of the cell. On the other hand, bundling proteins present in the cytoplasm such as a-actinin, form thick F-actin cables that transmit contractile forces along the cell during locomotion and help maintain the cell under a pre-stress state. Our group is developing a multi-scale approach for studying F-actin and ABP interactions ranging from network macromechanics to single molecule biophysics.
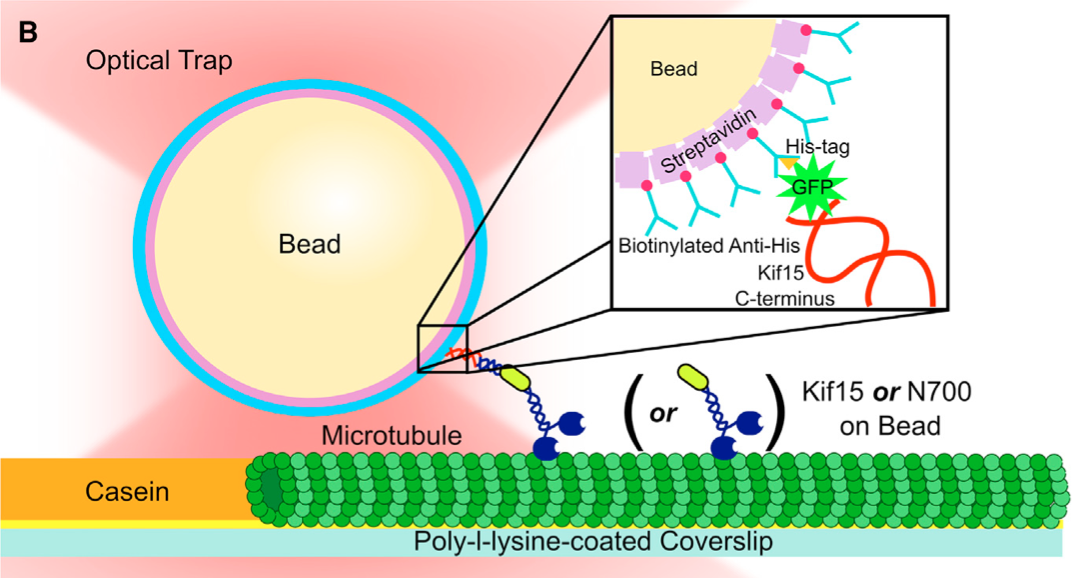
Bacteriophage Tethers
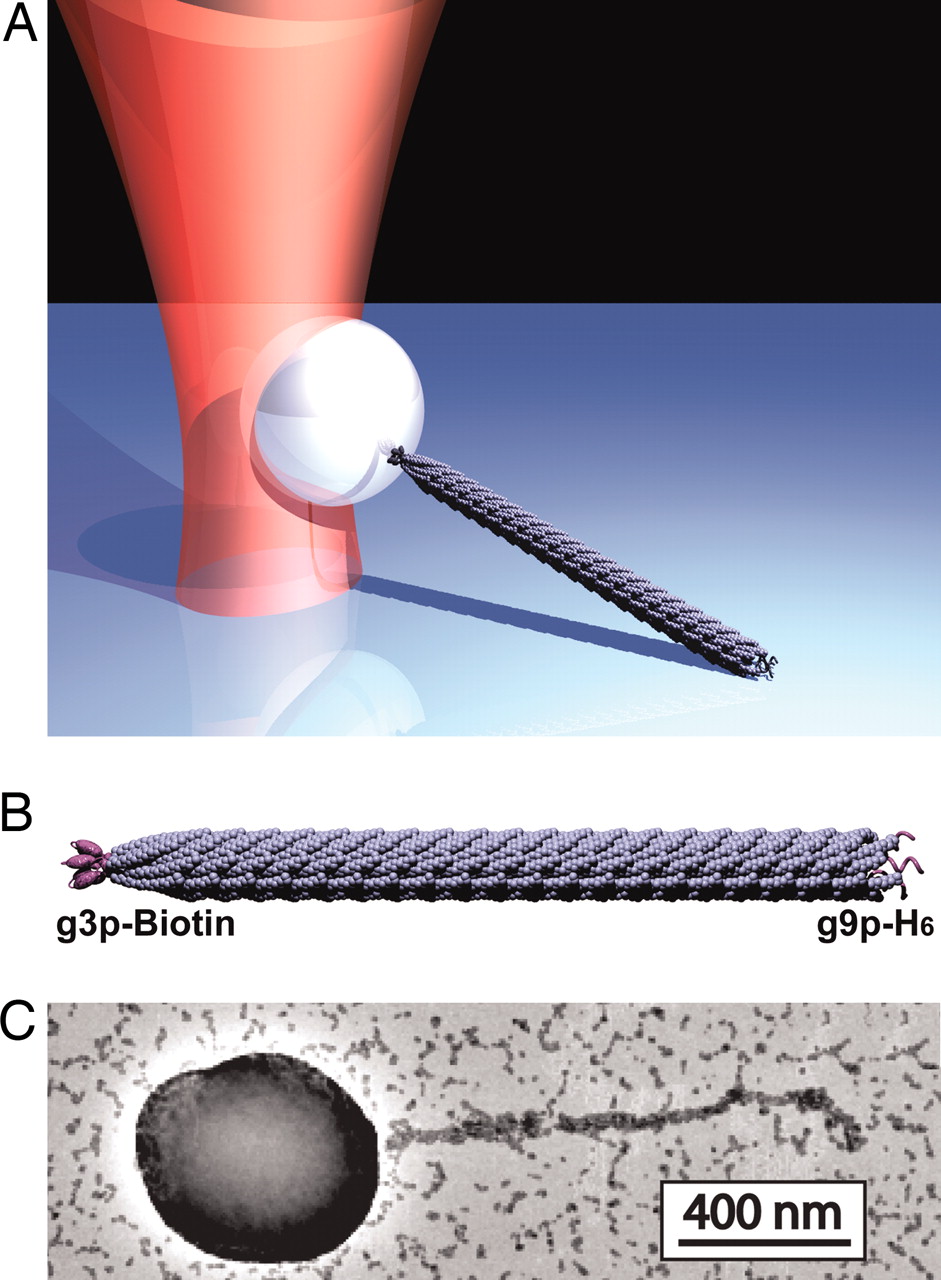
One of the greatest challenges in molecular biophysics is developing single molecule assays. Here we describe a new strategy for forming linkages between proteins of interest and an object, such as a bead, appropriate for use in the optical trap. In collaboration with Prof. Angela Belcher’s lab, we have developed a new strategy for forming these linkages based on the M13 bacteriophage. Our strategy uses genetic engineering to form the linkages, i.e., proteins of interest are covalently displayed on phage capsid proteins via genetic modification of the M13 genome. As a result, we exploit the natural assembly process of bacteriophages in E. coli to construct stand-alone tethers. We have characterized the mechanical properties of this tether and found that it has excellent properties: (1) Optimal length (~1um) for applying horizontal loads. In particular, it is long enough to calibrate and mitigate force correction terms, yet short enough to properly communicate mechanical transitions; (2) Stiffness 3X that of dsDNA; (3) Strength to withstand forces above 60pN. This work has been published in PNAS, see Khalil et al.