We are recruiting undergraduate researchers
Please contact chien.lun.hung@vanderbilt.edu with a statement of interest and resume
Signaling Proteins: Mechanism, Evolution, and Structure
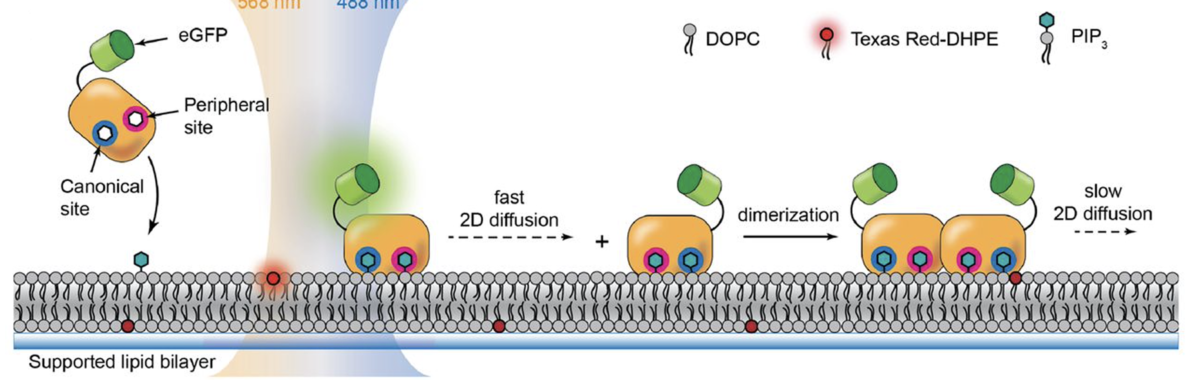
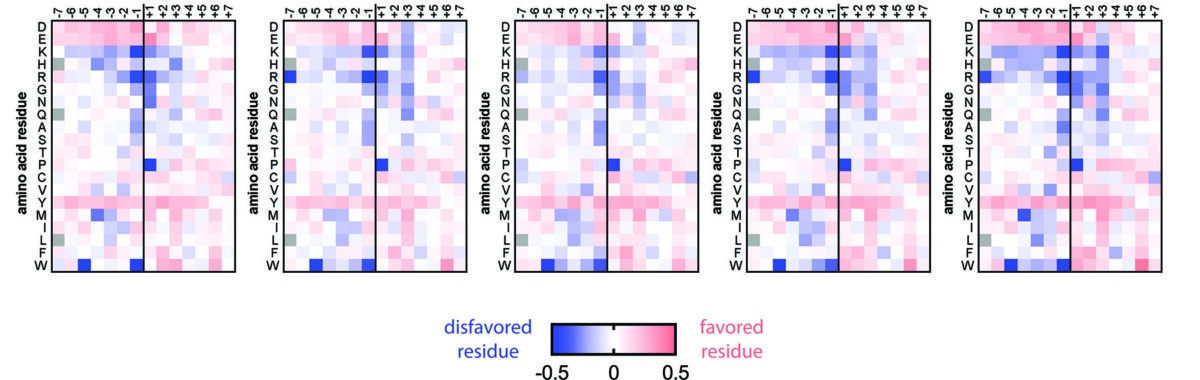
Our lab studies the mechanisms, evolution, and structures of the molecular switches that carry out cellular signal transduction. We use biochemical, biophysical, structural and cell biological analyses to elucidate mechanisms, and also study how these mechanisms change with evolution. A major focus of the lab is to understand the allosteric communication that enables proteins to be exquisitely responsive to input signals. We use high-throughput mutational analysis to determine the sensitivity of these mechanisms to perturbations, in order to determine the molecular principles governing regulation and specificity.
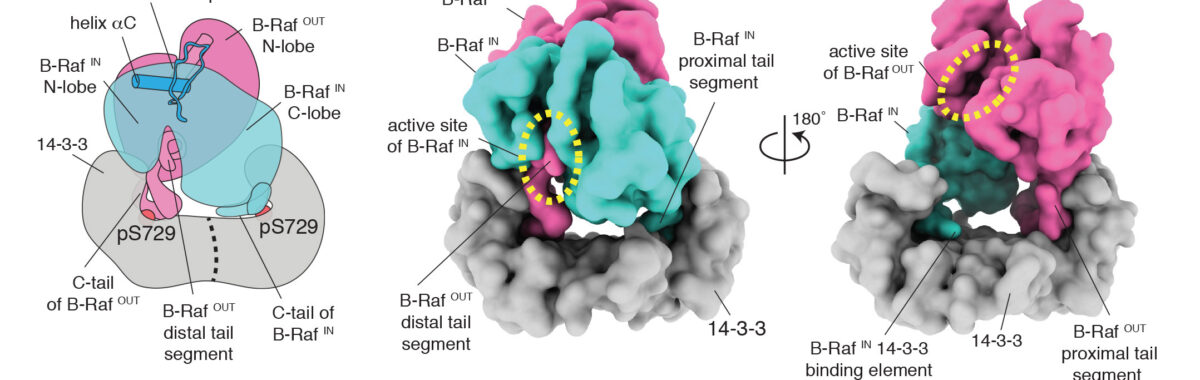
Breakthroughs from the lab have included the determination of the switching mechanisms of several tyrosine kinases, including that of the Src family of tyrosine kinases, the immune-cell kinases ZAP-70 and Btk, and the epidermal growth factor receptor (EGFR). We have also advanced the fundamental understanding of the regulation of several other signaling proteins, including the Ras activator SOS, and CaMKII, an oligomeric kinase that plays a central role in neuronal signaling. Our insights have helped understand how the misregulation of these enzymes is often coupled to cancer and immune diseases, and have implications for the development of kinase-targeted drugs to treat these diseases.
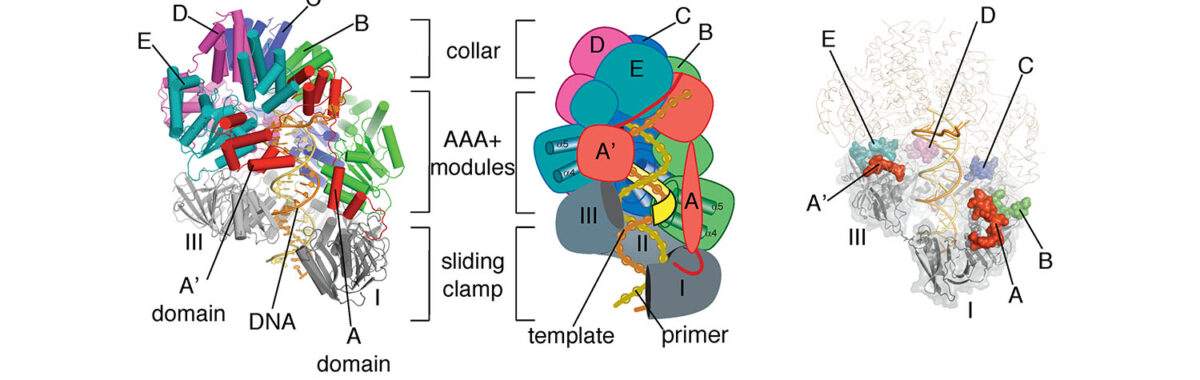
The lab has also made fundamental discoveries related to the structural basis for high-speed DNA replication, by determining the structures of the clamp-loader and sliding clamp complexes of DNA polymerases. We are currently applying high-throughput mutagenesis methods to these systems to understand the functioning of the AAA+ ATPases that form the core of the clamp-loader complexes.