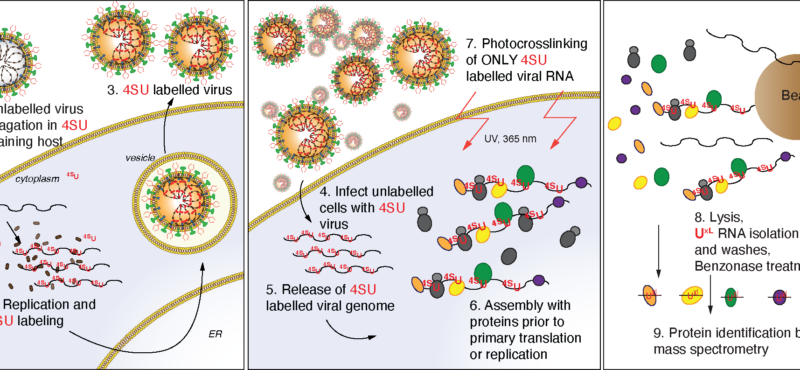
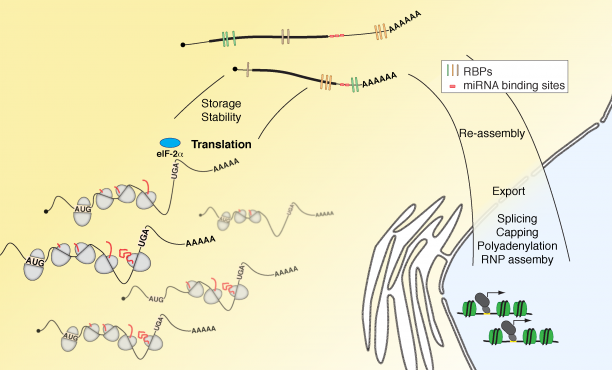
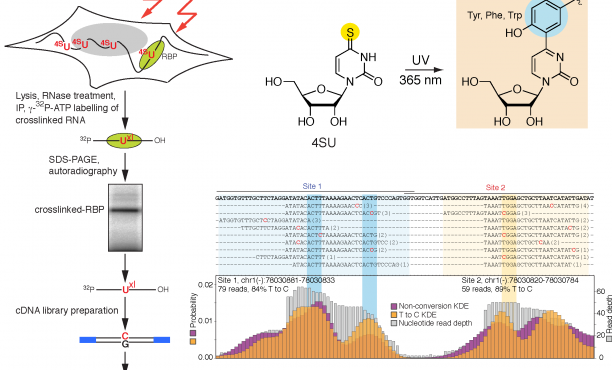
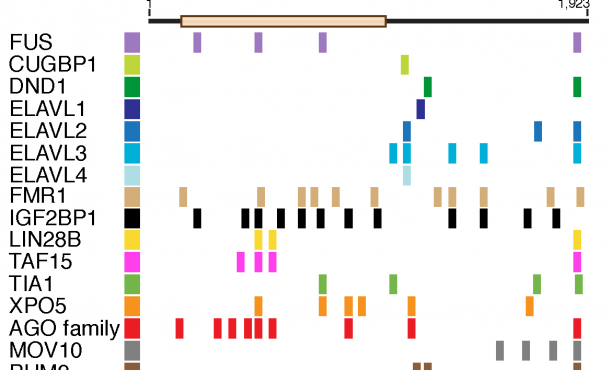
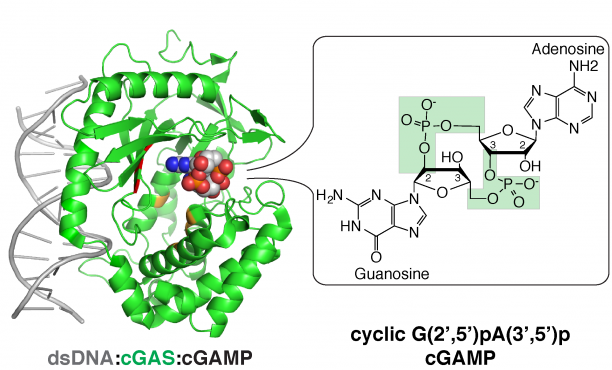
Aberrant RNA or DNA are robust triggers of the innate immune system as they can be early indicators of cellular damage or pathogenic infection by bacteria or virus. However, the criteria that could differentiate ‘self’ or ‘safe’ nucleic acids from ‘damaged’ or ‘foreign’ are not straightforward since the biochemical features of harmful RNA or DNA are not strictly unique to them. Viruses are particularly insidious as they have evolved strategies that either prevent their RNA or DNA genomes from being detected by pattern recognition receptors of the innate immune system and/or usurp specific machinery of cells in order to initiate viral replication. For example, owing to the rich tapestry of host RNA-binding proteins (RBPs) that facilitate cellular messenger RNA transcription, maturation, and translation, different RNA viruses have found ways to associate with cellular RBPs to exploit normal host RNA metabolic pathways in a manner that allows viral genomes to be processed, translated, and eventually replicated.
In an effort to understand the molecular arms race between host and pathogen, the research program of the Ascano laboratory is centered on exploring the mechanisms that cells use to differentiate whether a nucleic acid is derived from self or non-self, and the innate immune events that ensue following that decision point.
We have active research projects within two areas:
- The roles and coordination of RNA-binding proteins in regulating gene expression during innate immune activation or viral infection.
and
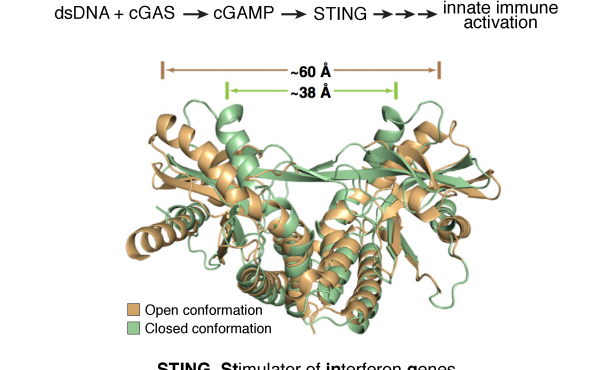
- The cytosolic DNA-sensing pathway involving the pattern recognition receptor and sensor cGAS, its second messenger product cGAMP, and the endoplasmic reticulum-bound receptor STING.
Our lab integrates novel biochemical, virological, immunological, and molecular and cell biological tools with high-throughput transcriptomic and proteomic technologies in an effort to elucidate the gene regulatory networks at play during immunologic stress. A deeper understanding of the physiological and pathological activities that surround RNA regulation enables us to leverage such information towards the development of novel vaccines and immunotherapeutics. The lab is affiliated with the Department of Biochemistry at the Vanderbilt University School of Medicine and is also part of the Microbe-Host Interactions Program within the Department of Pathology, Microbiology, and Immunology at VUMC.
We have multiple postdoctoral positions open:
RNA Regulatory Processes that govern Viral Host-Pathogen Interactions
About Us:
The Ascano Laboratory at the Department of Biochemistry, Vanderbilt University School of Medicine, is actively seeking a highly motivated and talented Postdoctoral Research Associate to join our dynamic team. Our research program is centered on unraveling the intricate molecular mechanisms that govern the interactions between host cells and pathogens, with a particular focus on RNA regulatory processes during viral infections.
Research Focus:
The successful candidate will have the opportunity to contribute to cutting-edge research in two key areas:
- Roles and Coordination of RNA-Binding Proteins:
- Investigate the functions of RNA-binding proteins in regulating gene expression during innate immune activation and viral infection.
- Explore how different RNA viruses associate with cellular RNA-binding proteins to exploit host RNA metabolic pathways.
- Virus Assembly and Extracellular Vesicle (EV) Assembly:
- Explore the relationship between virus and extracellular vesicle (EV) assembly, with a focus on RNA metabolic processes.
- Investigate how RNA regulatory pathways influence the assembly, release, and uptake of both viruses and EVs.
Qualifications:
We are seeking highly motivated individuals with a Ph.D. degree and a strong background in RNA biology and/or innate immunity, with preference given to those with computational experience. Our laboratory comprises an interdisciplinary group interested in examining RNA post-transcriptional mechanisms in innate immunity and host-viral pathogen interactions. We utilize biochemical, virological, immunological, and molecular and cell biological tools with high-throughput transcriptomic and proteomic technologies to answer fundamental questions on how gene expression is regulated at the RNA level during periods of innate immune activation and viral infection. We have funding available to support this postdoctoral position.
Benefits:
- Highly competitive salary with benefits.
- Lab and institution-level training resources for professional growth and development
- Access to new state-of-the-art facilities and resources.
- Highly collaborative and intellectually stimulating research environment.
Application Process:
Interested candidates should submit a CV, a cover letter detailing research interests and accomplishments, and contact information for three references to manuel.ascano@vanderbilt.edu. We encourage candidates from diverse backgrounds to apply. Vanderbilt University is an equal opportunity employer.
Affiliation:
The Ascano Laboratory is affiliated with the Departments of Biochemistry and Pathology, Microbiology, and Immunology at Vanderbilt University. We are also members of the Center for Immunobiology, Center for Extracellular Vesicle Research, Institute for Infection, Immunology, and Inflammation, and Institute for Chemical Biology.
About Nashville:
Embark on your postdoctoral journey in Nashville, a vibrant city renowned for its thriving music scene, diverse cultural offerings, and dynamic scientific community. As emerging scientists in the early stages of their careers, you’ll find Nashville to be a hub of innovation and creativity. Beyond the lab, immerse yourself in the city’s progressive atmosphere, where Nashville’s unique character complements its rich cultural tapestry. Explore a city that allows you to conduct groundbreaking research with a young and progressive community, making your postdoctoral experience truly exceptional.
As part of the Department of Biochemistry in the School of Medicine – Basic Sciences, our lab has access to state-of-the art facilities to enable scientific breakthroughs. At Vanderbilt University, we are a community of learning committed to diversity, equity, and inclusion.
Join us!
Long-term funding is secure, along with a competitive salary and benefits. Highly accomplished applicants may also be eligible for Destination Biochemistry Postdoctoral Scholar competitive awards.
Interested individuals should contact PI directly for more information.