Software
EnzyHTP: A High-Throughput Computational Platform for Enzyme Modeling
Sep. 21, 2025—EnzyHTP is a holistic platform that allows high-throughput molecular simulation of enzymes. The large-scale collection of molecular simulation data presents a significant challenge due to complex demands. To build an enzyme model appropriate for simulations, multiple hierarchies of structural definitions and treatments must be established such as protein stoichiometry, binding site, predicting amino acid protonation...
Entropic Path Sampling
Sep. 21, 2025—Entropic path sampling (EPS) was developed by our group as a trajectory-based protocol to quantify how configurational entropy varies along a reaction path, especially after the transition state, by harvesting ensembles of snapshots from quasiclassical direct-dynamics trajectories, slicing them into structural windows (e.g., by forming-bond length), and computing entropy with an internal-coordinate, histogram-based estimator using...
Deep Learning Models for Protein Function Prediction
Sep. 21, 2025—EnzyKR and DeepLasso illustrate how task-specific neural architectures, grounded in physical constraints (activation barriers, topology) and trained on structure- or library-scale data, can deliver actionable predictions for enzyme selectivity and RiPP bioactivity in real design workflows. EnzyKR. EnzyKR is a chirality-aware deep learning framework for stereoselective biocatalysis. It uses a classifier–regressor pipeline that first identifies...
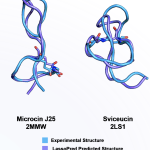
LassoHTP/LassoPred for Lasso Peptide Structure Prediction and Modeling
Sep. 20, 2025—Lasso peptides (LaPs) are RiPPs with a characteristic slipknot (lariat-knot) topology that confers exceptional thermal stability, protease resistance, and often potent bioactivity. Despite thousands of LaP sequences identified by bioinformatics, only ~50 have experimentally determined structures, largely because their irregular scaffold and isopeptide bond confound general-purpose predictors. LassoPred/LassoHTP enables end-to-end prediction and modeling of LaPs...
Information Extraction and Database Construction
Jan. 24, 2021—The Yang lab is building an integrated enzymology data ecosystem that makes the “dark matter” of enzyme kinetics accessible for predictive modeling and method development. IntEnzyDB provides a fast, flattened relational architecture that unifies enzyme structure and function across six EC classes and exposes a public web interface for streamlined access; using 1,050 structure-kinetics pairs,...