EnzyHTP: A High-Throughput Computational Platform for Enzyme Modeling
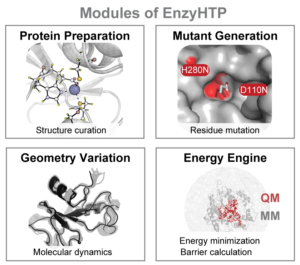
EnzyHTP is a holistic platform that allows high-throughput molecular simulation of enzymes. The large-scale collection of molecular simulation data presents a significant challenge due to complex demands. To build an enzyme model appropriate for simulations, multiple hierarchies of structural definitions and treatments must be established such as protein stoichiometry, binding site, predicting amino acid protonation state, addition of missing residues, performing an amino acid substitution, and creating reacting species. Most enzyme modeling practices use similar structural operations but rely on manual curation, which is highly inefficient and hampers reproducibility. EnzyHTP, a high-throughput enzyme simulation tool, bypasses these issues through automation of molecular model construction, mutation, sampling, and energy calculation.
Qianzhen Shao is leading our development team of EnzyHTP. The team is actively working on new features as well as refactoring EnzyHTP to make it more user-friendly and increase the extensibility for future incorporation of more functions.
The software code:
https://github.com/ChemBioHTP/EnzyHTP
Tutorials:
https://enzyhtp-doc.readthedocs.io/en/latest/
Publications of EnzyHTP:
https://pubs.acs.org/doi/10.1021/acs.jcim.1c01424
https://pubs.acs.org/doi/10.1021/acs.jcim.3c00618
Leave a Response
You must be logged in to post a comment