YcaO chemistry
In the hunt for improved antibiotics, it is important to realize that natural products have been, without question, the most prolific source of all medicines. With the advent of massively parallel DNA sequencing, it has become apparent that our knowledge of natural product structure and function is astonishingly incomplete. Exploration of uncharted natural product chemical space will undoubtedly lead to improved, and entirely new, medicines. Thus, our group focuses on elucidating the biosynthesis, structure, and function of natural products. Our primary focus has been on thiazole/oxazole-containing peptides. Characterized examples have activities ranging from antibacterials to virulence-promoting toxins. Therefore, the study of these peptidic natural products allows us to not only better understand bacterial virulence (where pharmacological intervention would constitute a pathogen-specific approach to bacterial infection) but also explore unique chemical architectures, ideally positioning us to introduce new structural classes of antibiotics. Our recent successes and focal areas are highlighted below by research project.
We are studying the structural and chemical nature of a unique class of biosynthetic enzymes.
Recent Publications
Liu, A.; Si, Y.; Dong, S.; Mahanta, N.; Penkala, H.N.; Nair, S.K.; Mitchell, D.A. “Functional elucidation of TfuA in peptide backbone thioamidation.” Nat. Chem. Biol. (2021). doi:https://doi.org/10.1038/s41589-021-00771-0
Nayak, D.D.; Liu, A.; Agrawal, N.; Rodriguez-Carerro, R.; Dong, S.H.; Mitchell, D.A.; Nair, S.K.; Metcalf, W.W. “Functional interactions between posttranslationally modified amino acids of methyl-coenzyme M reductase in Methanosarcina acetivorans.” PLoS Biol. (2020). doi:https://doi.org/10.1371/journal.pbio.3000507
Dong, S.H.; Liu, A.; Mahanta, N.; Mitchell, D.A., Nair, S.K. “Mechanistic basis for ribosomal peptide backbone modifications.” ACS Cent. Sci., 5: 842-851 (2019). doi:10.1021/acscentsci.9b00124
Mahanta, N.; Szantai-Kis, D.M.; Petersson, E.J.; Mitchell, D.A. “Biosynthesis and chemical applications of thioamides.” ACS Chem. Biol., 14: 142-163 (2019). doi:10.1021/acschembio.8b01022
The defining chemical feature of the linear azole-containing peptide (LAP) family of natural products are thiazol(in)e and (methyl)oxazol(in)e heterocycles, which derive from cysteine and serine/threonine residues of a ribosomally produced precursor peptide. These heterocycles also appear ubiquitously in the thiopeptides as well as in certain cyanobactins . Upon heterocyclization, an otherwise unstructured peptide becomes rigidified and able to elicit a biological function. It is common for numerous residues of the precursor peptide to undergo heterocyclization, among other posttranslational modifications, to yield the mature natural product. Our group investigated azoline/azole biosynthesis given that they are found in many pharmacologically active compounds and, prior to our work, the details of their formation were poorly understood.
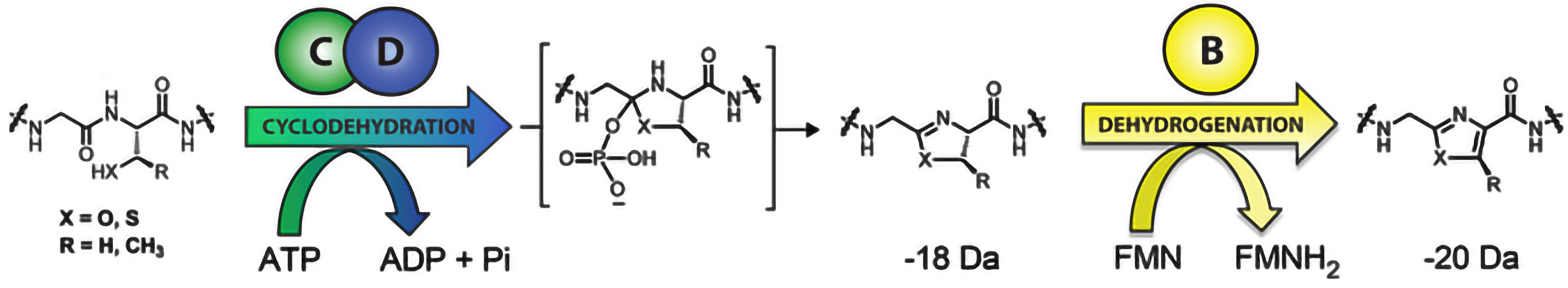
In a two-step sequence, an ATP-dependent cyclodehydration first forms the azoline heterocycle, followed by an optional FMN-dependent dehydrogenation to afford the aromatic azole. (catalyzed by C/D) first forms the azoline heterocycle, followed by an optional FMN-dependent dehydrogenation (catalyzed by B) to afford the aromatic azole. Past attempts to gain deeper insights into the mechanism of heterocyclization and the specificity of the enzyme complex has been stymied by poor protein solubility, stability and difficulties monitoring heterocycle formation. Bioinformatically, we identified linear azol(in)e-containing peptide clusters that did not suffer from the same pitfalls, allowing us to answer previously intractable questions regarding the mechanistic details of linear azol(in)e-containing peptide biosynthesis. These investigations deciphered the regio- and chemoselectivity of substrate processing by a rather promiscuous cyclodehydratase and established the mechanistic role of ATP hydrolysis in the cyclodehydration reaction. This work revealed a new biological role for ATP, which is used to directly phosphorylate peptide backbone oxygens. Moreover, we have shown by NMR and other biochemical assays that it is actually the D-protein component, and not the C-protein, that performs the cyclodehydration reaction. The function of these collaborating proteins was heretofore not dissectible.
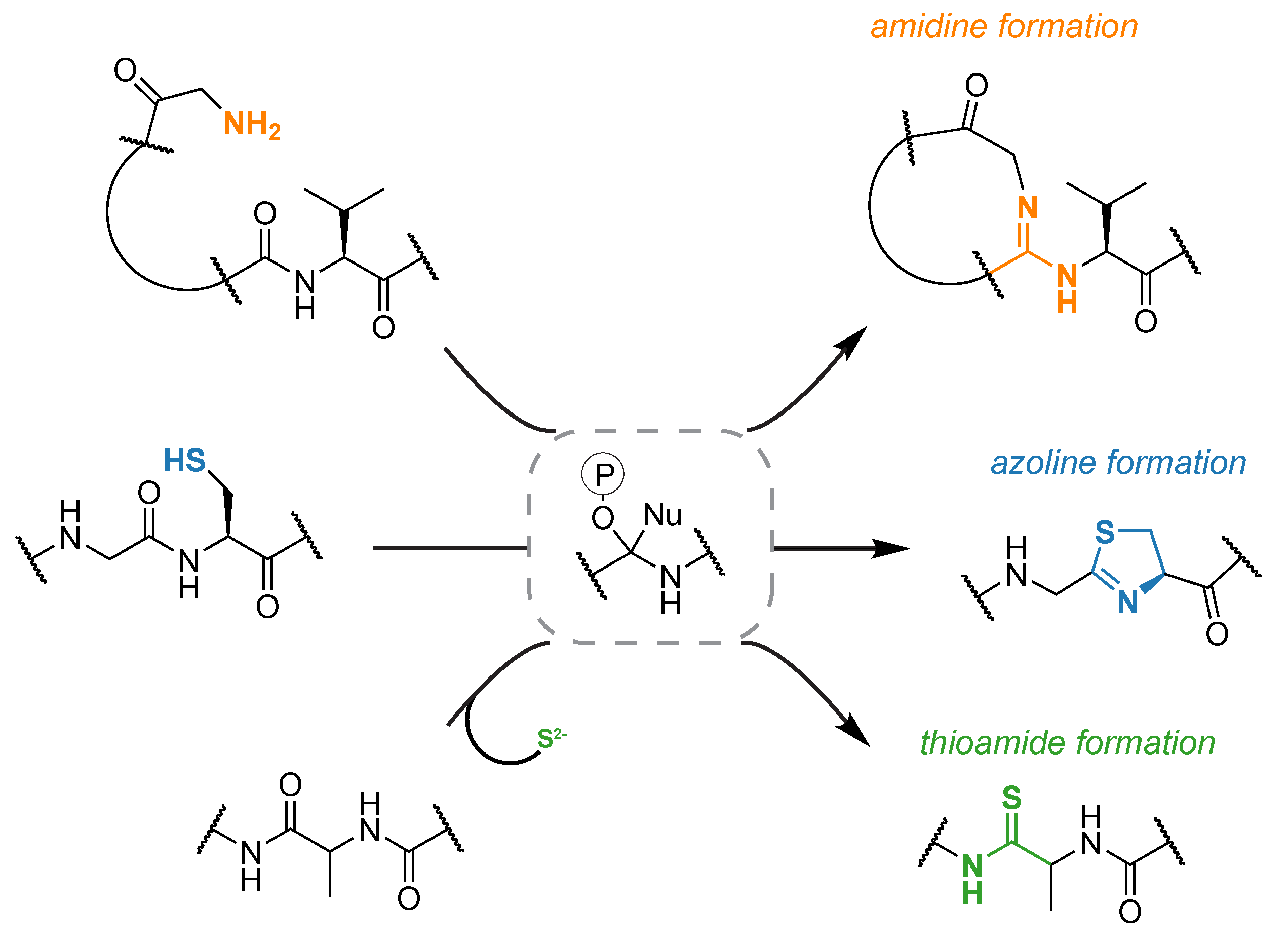
More recently we have been investigating divergent members of the enzyme family that includes the cyclodehydratase shown above. Collectively, these proteins are referred to as YcaO enzymes but only about involved in azoline biosynthesis. Genome neighborhood analysis predicts that out of all currently identified YcaO proteins (n=14,000) less than one-third are encoded in a context that would allow for azoline formation. Approximately 10% of the total are encoded next to a gene named tfuA, but the predicted precursor peptide often lack the plethora of beta-nucleophile containing residues required for heterocycle formation. We have found that these biosynthetic gene clusters are responsible for peptidic thioamidation and require sulfur from an outside source. Interestingly, methanogenic archaea always encode a YcaO protein. We have recently found that the YcaO is directly responsible for the thioamidation of methyl-coenzyme M reductase, one of the most important proteins in the global carbon cycle. YcaO proteins are also responsible for the formation of the macrolactamidine functional group found in bottromycin. In this example, the N-terminus of the peptide, as opposed to an adjacent Cys/Ser/Thr or exogenous sulfide, is the nucleophile that attacks the activate amide backbone. Nature clearly has employed YcaO proteins widely to perform chemistry on the backbone of peptides and we anticipate new reactions to be discovered within and beyond this enzyme family.