Using Bioinformatics to Aid Discovery: RODEO
Natural products, or specialized molecules produced by living things, have historically been an important source of antibiotics and other drugs. To accelerate the discovery of new natural products, researchers have turned to bioinformatics methods for predicting whether a specific organism will make an undiscovered molecule. 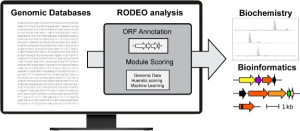 This has led to an increase in “genome mining” for natural product biosynthetic pathways. While genome mining can access diverse natural products, the process can be time consuming and the number of organisms with sequenced genomes continues to explode. As a result, automation of genome mining and big data analysis pipelines is crucial for the prioritization and discovery of new natural products.
This has led to an increase in “genome mining” for natural product biosynthetic pathways. While genome mining can access diverse natural products, the process can be time consuming and the number of organisms with sequenced genomes continues to explode. As a result, automation of genome mining and big data analysis pipelines is crucial for the prioritization and discovery of new natural products.
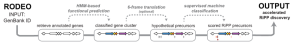
To lead these efforts, the Mitchell Lab has developed an algorithm called RODEO, which aids genome miners by automating the annotation and visualization of genomic data. While RODEO is a multi-purpose algorithm, it specifically includes modules designed to analyze certain types of RiPPs (ribosomally synthesized and post-translationally modified peptides) using motif analysis, heuristic scoring, and supervised machine learning. RiPPs offer an exciting opportunity for natural product discovery, as peptides and other biologics are increasingly sought-after therapeutic modalities. By studying RiPP biosynthesis, we hope to access new bioactive molecules as pharmaceutical leads and new enzymes as tools for biotechnological or pharmaceutical development. The Mitchell lab maintains RODEO as a public webtool, and RODEO is under active development to further aid researchers studying RiPPs.
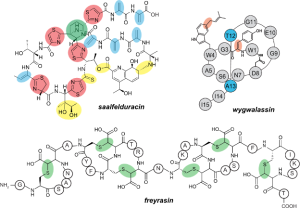
Beyond bioinformatic methods, we also validate and characterize new natural products predicted by RODEO. Following dataset generation and analysis, we select cases with the potential for discovering new natural products and/or enzymatic transformations. Beginning with our original report of RODEO in 2017, we have expanded molecular diversity in many RiPP classes including lasso peptides, graspetides, thiopeptides, lanthipeptides, linaridins, and borosins, and discovered new RiPP classes defined by new biosynthetic reactions such as the ranthipeptides, daptides, and aminopyruvatide. RODEO also allows us to explore the larger evolutionary contexts of RiPP biosynthesis, from both computational and biochemical perspectives. At the moment, our efforts with RODEO are focused on pushing the boundaries of hypothesis generation using bioinformatics. Collectively, these approaches are enabling multiple lines of investigation for the discovery of new RiPPs, study of their biological functions, and the engineering of RiPP biosynthetic systems for biotechnological applications.
Key Publications:
3. A scalable platform to discover antimicrobials of ribosomal origin. Nat. Commun., 13: 6135 (2022).