Thiopeptides
In the hunt for improved antibiotics, it is important to realize that natural products have been, without question, the most prolific source of all medicines. With the advent of massively parallel DNA sequencing, it has become apparent that our knowledge of natural product structure and function is astonishingly incomplete. Exploration of uncharted natural product chemical space will undoubtedly lead to improved, and entirely new, medicines. Thus, our group focuses on elucidating the biosynthesis, structure, and function of natural products. Our primary focus has been on thiazole/oxazole-containing peptides. Characterized examples have activities ranging from antibacterials to virulence-promoting toxins. Therefore, the study of these peptidic natural products allows us to not only better understand bacterial virulence (where pharmacological intervention would constitute a pathogen-specific approach to bacterial infection) but also explore unique chemical architectures, ideally positioning us to introduce new structural classes of antibiotics. Our recent successes and focal areas are highlighted below by research project.
We are studying the biosynthesis of thiopeptide natural products.
Recent Publications
Nguyen, D.T.; Le, T.T.; Rice, A.J.; Hudson, G.A.; van der Donk, W.A.; Mitchell, D.A. “Accessing Diverse Pyridine-Based Macrocyclic Peptides by a Two-Site Recognition Pathway” J. Am. Chem. Soc. (2022). doi:https://doi.org/10.1021/jacs.2c02824
Hudson, G.A.; Hooper, A.R.; DiCaprio, A.J.; Sarlah, D.; Mitchell, D.A. “Structure prediction and synthesis of pyridine-based macrocyclic peptide natural products.” Org. Lett. (2020). doi:https://doi.org/10.1021/acs.orglett.0c02699
Schwalen, C.J., Hudson, G.A., et al. “Bioinformatic Expansion and Discovery of Thiopeptide Antibiotics.” J. Am. Chem. Soc. (2018)
doi:10.1021/jacs.8b03896
Cogan, D.P., Hudson, G.A., et al. “Structural Insights into Enzymatic [4+2]-Aza-cycloaddition in Thiopeptide Antibiotic Biosynthesis.” Proc. Natl. Acad. Sci. (2017)
doi:10.1073/pnas.1716035114
The chemical hallmark of the linear azole-containing peptide (LAP) family of natural products are thiazol(in)e and (methyl)oxazol(in)e heterocycles installed by cyclodehydratases from cysteine and serine/threonine residues, respectively. These post-translational modifications impart rigidity on a peptidic natural product and in many cases are crucial for downstream enzymatic processing and bioactivity. Reported in a series of papers from 2012-2015, we thoroughly investigated the details of cyclodehydration, including the mechanistic enzymology of azoline biosynthesis, which uses ATP to directly activate the peptidic amide backbone, the structural details of the cyclodehydratase, revealing a novel ATP binding motif, and the orchestration of the flavin-dependent dehydrogenase which oxidizes azolines to corresponding azoles. Further research revealed a new class of cyclodehydratases which, upon characterizing, deciphered how the peptidic substrate is recruited to the cyclodehydratase at the molecular level.
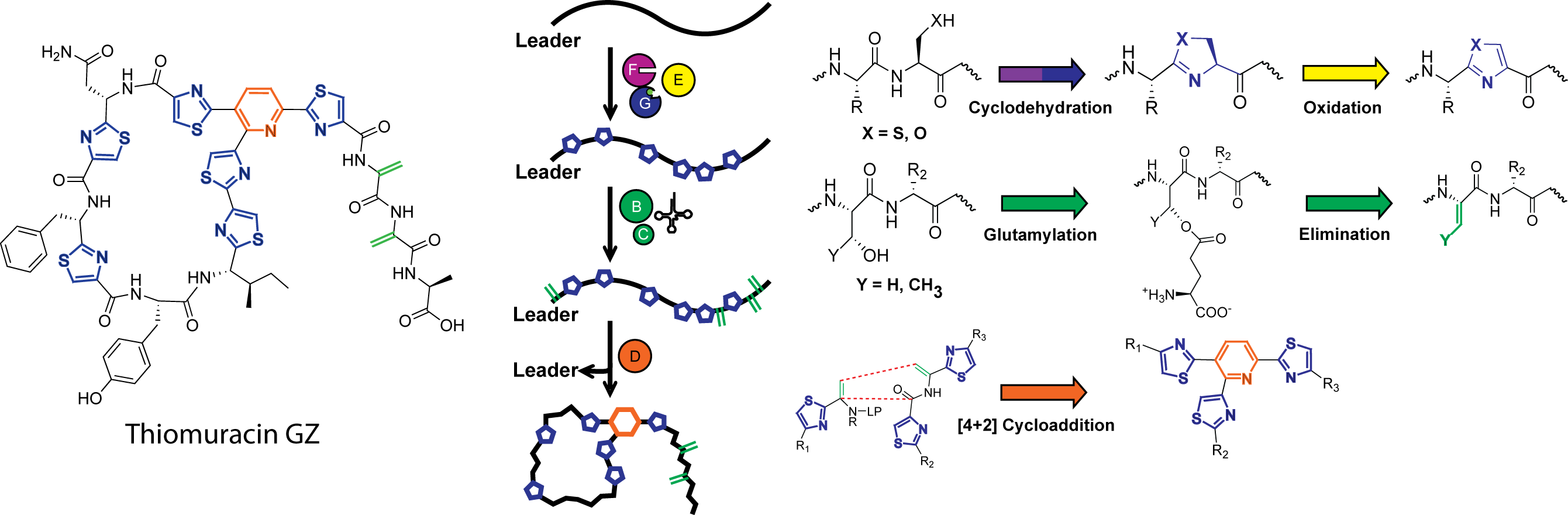
Gaining a deeper understanding of cyclodehydration has enabled the study of more complicated systems, including the thiopeptides, a family of peptidic natural products with potent antibiotic activity towards a variety of drug-resistant bacteria. Despite their architectural complexity, thiopeptide biosynthesis proceeds through a LAP-like intermediate, and as such, feature cyclodehydratases and dehydrogenases for thiazole biosynthesis, but also feature lanthipeptide-like dehydratases and a [4+2] cycloaddition enzyme responsible for pyridine formation. Building upon our previous work on cyclodehydratases, and in collaboration with lanthipeptide biosynthetic experts in the laboratory of Prof. Wilfred van der Donk, we have reconstituted the core biosynthesis of thiomuracin, enabling production of these complicated molecules in vitro using only six enzymes on a precursor peptide substrate. Reconstitution also provided unprecedented insight into the precise details of thiopeptide biosynthesis, including the largely uncharacterized [4+2] cycloaddition enzyme. Further endeavors have yielded crucial insight into the timing and substrate specificity of thiomuracin biosynthesis and will provide a groundwork for enzymatically generating new thiopeptide analogs with desirable pharmacological properties.