Research Overview
In the hunt for improved antibiotics, it is important to realize that natural products have been, without question, the most prolific source of all medicines. With the advent of massively parallel DNA sequencing, it has become apparent that our knowledge of natural product structure and function is astonishingly incomplete. Exploration of uncharted natural product chemical space will undoubtedly lead to improved, and entirely new, medicines. Thus, our group focuses on elucidating the biosynthesis, structure, and function of natural products. Our primary focus has been on thiazole/oxazole-containing peptides. Characterized examples have activities ranging from antibacterials to virulence-promoting toxins. Therefore, the study of these peptidic natural products allows us to not only better understand bacterial virulence (where pharmacological intervention would constitute a pathogen-specific approach to bacterial infection) but also explore unique chemical architectures, ideally positioning us to introduce new structural classes of antibiotics. Our recent successes and focal areas are highlighted below by research project.
Lasso Peptides
|
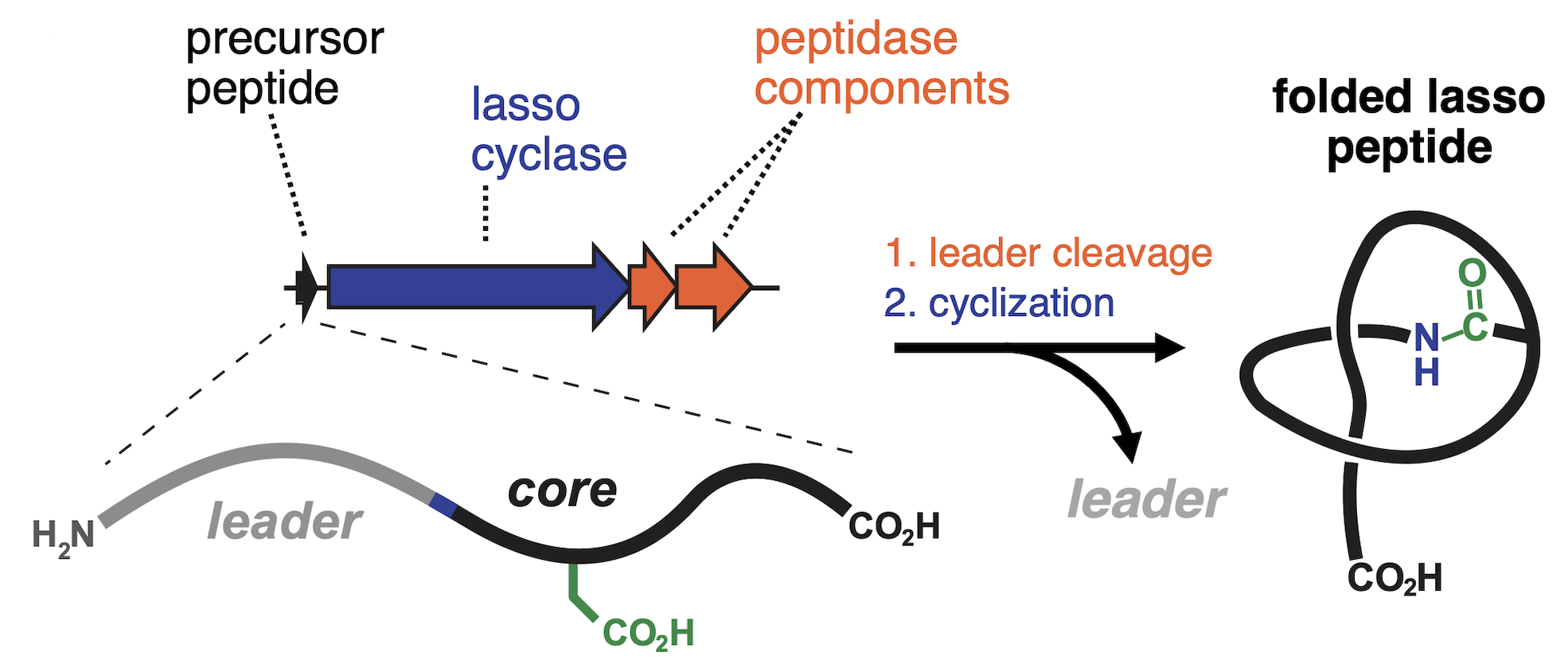
Lasso peptides are ribosomally synthesized natural products distinguished by a unique knotted topology that confers exceptional stability and protease resistance. The Mitchell Lab has pioneered genome-wide discovery of lasso peptides, revealing thousands of previously unrecognized systems and dramatically expanding the known sequence space of this family. Through a combination of bioinformatics, biochemical reconstitution, and enzyme engineering, we have uncovered remarkable biosynthetic flexibility that enables the creation of customized, non-natural lasso peptides. These efforts position lasso peptides as versatile scaffolds for molecular engineering and therapeutic development.
|
|
YcaO Enzymes
|
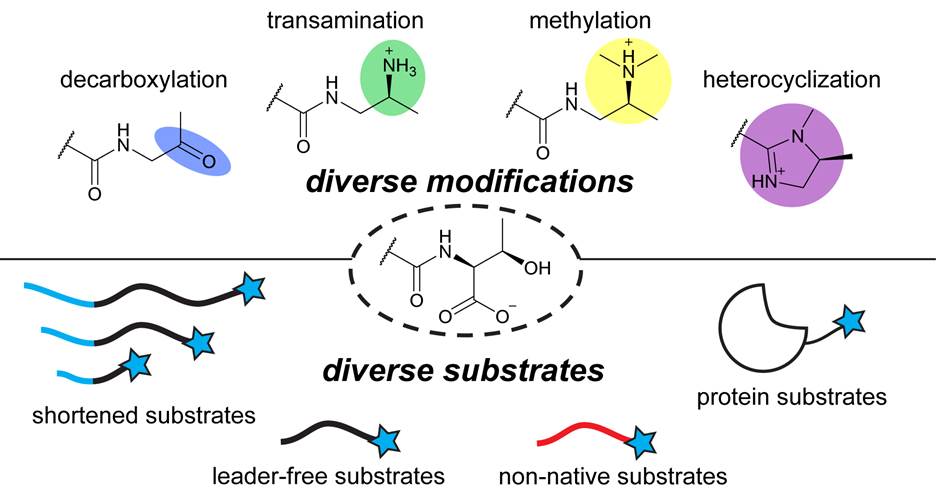
Our lab investigates YcaO enzymes, a widespread and versatile family of peptide-modifying enzymes that catalyze chemically unusual transformations on peptide backbones. By combining structural biology, biochemistry, and genetics, we have shown that YcaOs function as main-chain-modifying kinases capable of installing diverse modifications, including heterocycles and thioamides. These seemingly subtle chemical changes can profoundly alter peptide structure, stability, and biological function. With tens of thousands of YcaO homologs encoded across microbial genomes, defining their functions and biological roles remains a central focus of the lab.
|
|
RiPP Display
|
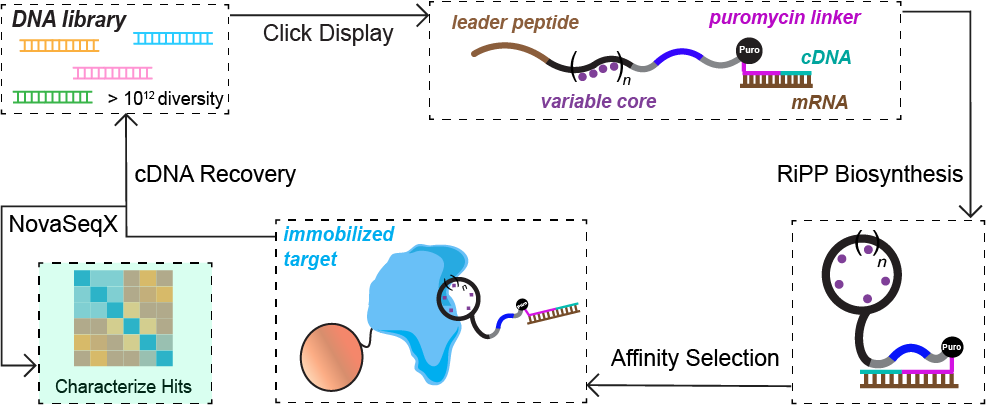
Ribosomally synthesized and post-translationally modified peptides (RiPPs) offer a rich and largely untapped source of structurally constrained scaffolds for drug discovery. Building on the growing success of peptide therapeutics, the Mitchell Lab is adapting high-throughput display technologies to evolve RiPP-derived molecules against therapeutically relevant targets. By linking peptide biosynthesis to large combinatorial libraries, these approaches enable the discovery of new bioactive compounds while simultaneously probing enzyme substrate tolerance at unprecedented scale. This strategy bridges natural product biosynthesis with modern directed evolution to explore previously undruggable biological space.
|
|
RiPP Recognition Element
|
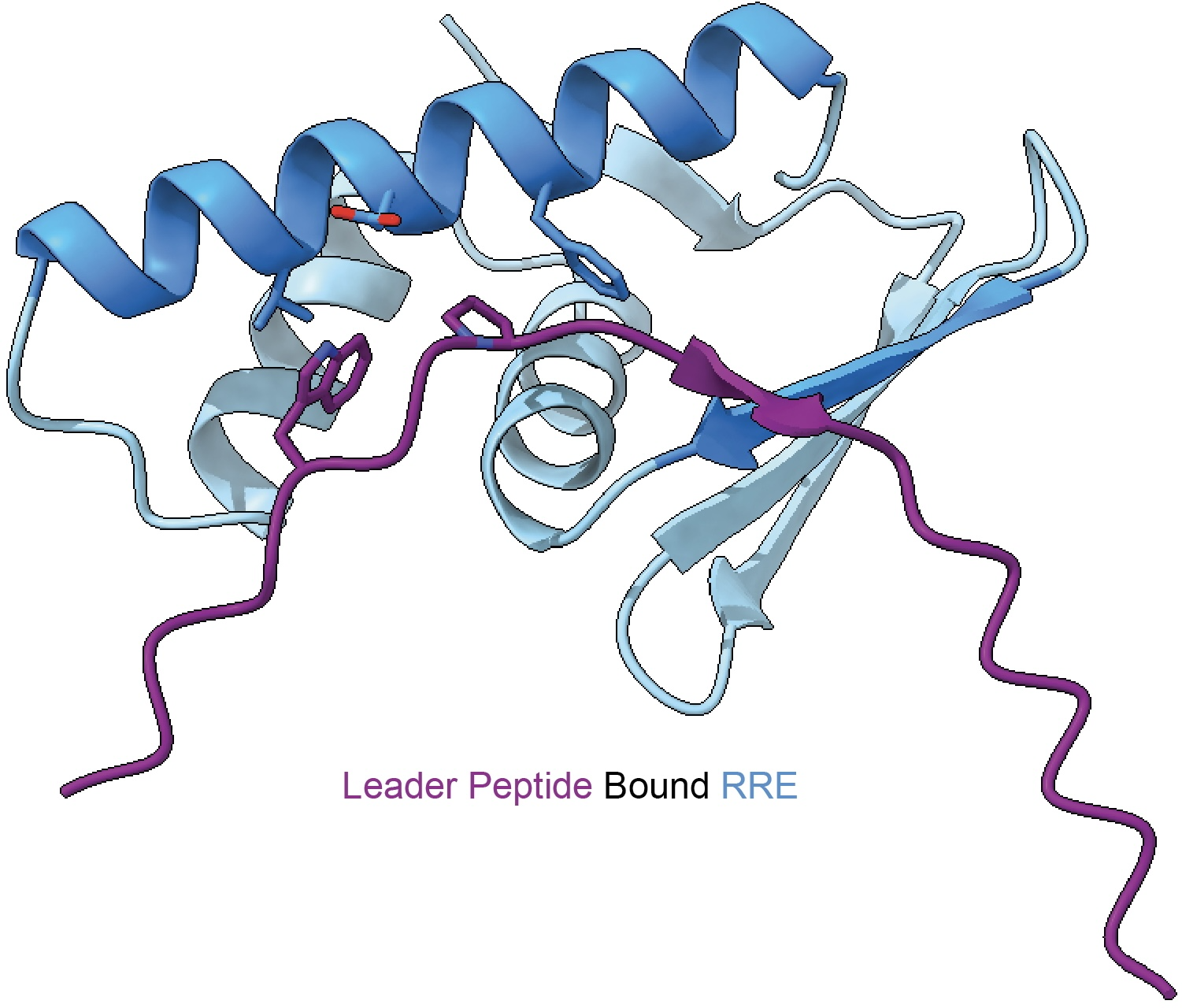
The RiPP Recognition Element (RRE) is a peptide-binding domain that plays a central role in directing post-translational modification during RiPP biosynthesis. Our work established the RRE as a widespread and unifying feature across diverse RiPP classes, despite minimal sequence similarity. By elucidating how RREs recognize precursor peptides and activate biosynthetic enzymes, we have uncovered fundamental principles governing RiPP assembly. Leveraging these insights, we use RRE-guided genome mining to discover entirely new RiPP pathways and enzymatic chemistries.
|
|
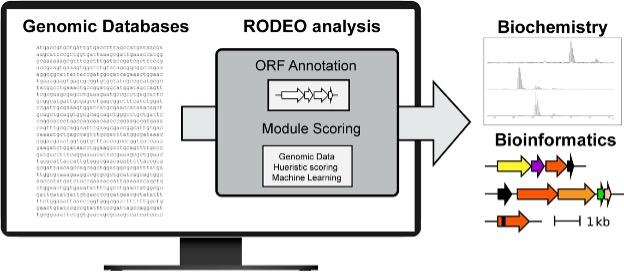
RODEO
|
To accelerate natural product discovery in the era of large-scale genome sequencing, the Mitchell Lab developed RODEO, a bioinformatics platform that automates genome mining and prioritization of ribosomally synthesized natural products. RODEO integrates motif analysis, heuristic scoring, and machine learning to identify and visualize biosynthetic gene clusters across diverse RiPP classes. Beyond prediction, our lab experimentally validates and characterizes new molecules and enzymes uncovered by these analyses. Together, these efforts combine computation and experiment to systematically expand natural product diversity and uncover new biochemical transformations.
|
|




