Lasso Peptides
Lasso peptides are defined by a uniquely knotted topology in which the C-terminus is threaded through an N-terminal macrolactam ring. This topology confers exceptional structural rigidity and protease resistance, making lasso peptides attractive scaffolds for molecular engineering. To facilitate systematic discovery of lasso peptides from genomic data, we developed Rapid ORF Description and Evaluation Online (RODEO) to automate the time-consuming process of annotating biosynthetic gene clusters and predicting likely lasso peptide precursors. Using RODEO, we performed the first comprehensive mapping of lasso peptide biosynthetic gene clusters across bacterial genomes, revealing thousands of previously unrecognized systems and dramatically expanding known lasso peptide sequence space. This large-scale analysis uncovered extensive diversity in macrolactam ring size, tail length, and steric plug composition, fundamentally redefining the biosynthetic landscape of the lasso peptide family and providing a global framework for understanding their natural variation.
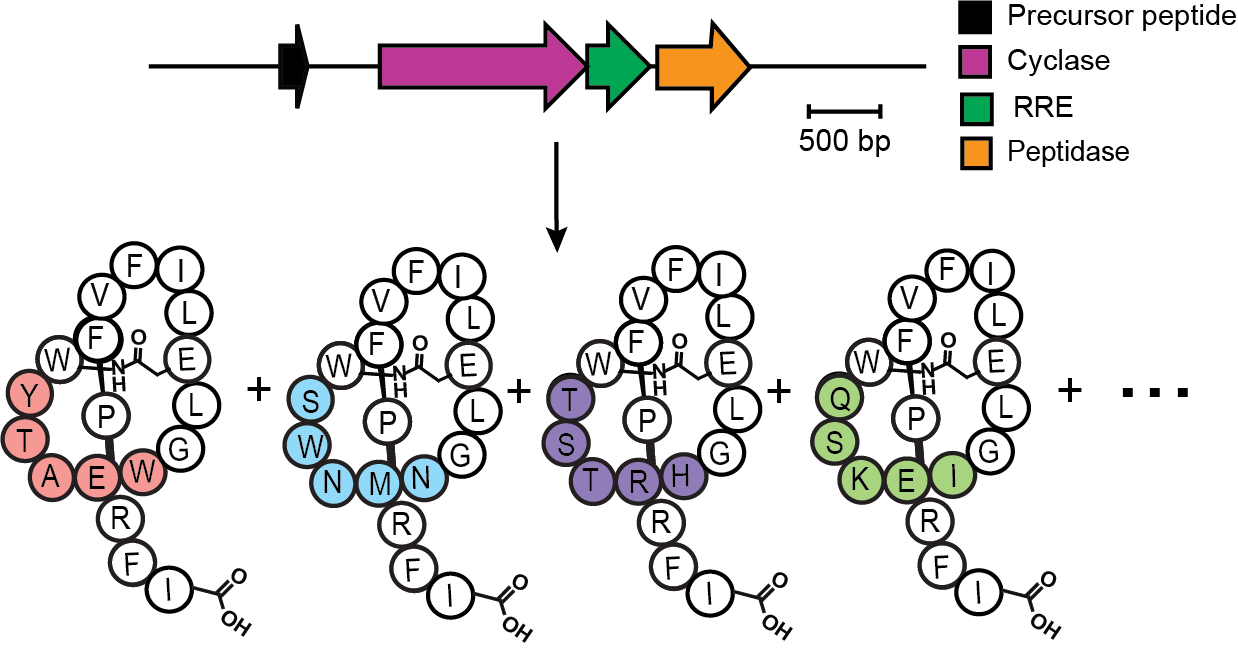
In 2019, RODEO enabled the discovery and characterization of fusilassin. This thermophilic lasso peptide system is only the second to date compatible with in vitro experiments, opening the doors for in-depth biochemical characterization of its enigmatic biosynthetic enzymes. Using cell-free technology, we were able to show that Fusilassin’s biosynthetic enzymes are extremely promiscuous, tolerating ~2 million diverse ring sequences while still producing correctly folded lasso peptides. These results revealed an unexpected degree of enzymatic promiscuity and established that lasso cyclases can operate across vast regions of sequence space. Together, the combination of large-scale bioinformatic discovery and biochemical reconstitution has positioned lasso peptides as one of the most versatile RiPP scaffolds known. Despite these advances, fundamental questions remain regarding how cyclase enzymes recognize substrates, enforce threading directionality, and prevent unthreading during catalysis. Ongoing work in the lab focuses on elucidating these mechanistic principles and defining the sequence–structure relationships that govern successful lasso peptide formation.

In parallel to bioinformatic discovery efforts, we are exploring lasso peptides as scaffolds for therapeutic design. Their compact size, rigid topology, and intrinsic protease resistance make them attractive candidates for targeting challenging protein–protein interactions and cell surface receptors. By integrating genome mining, enzyme engineering, and high-throughput screening strategies, we aim to access new-to-nature lasso peptide architectures with tailored chemical functionality and improved pharmacological properties, bridging fundamental biosynthetic insight with translational potential.
Key Publications: