Lab Highlights
| Shravan and collaborators from the Pamer lab have published an article in Cell Host & Microbe outlining the relationships between lanthipeptide-producing gut microbes and their effects on microbiome structure. Metagenomic data was collected and partially assembled from more than 1,000 clinical fecal samples. Using RODEO, Shravan searched these metagenomes for lanthipeptide biosynthetic gene clusters, and collaborators used this information to draw correlations between lanthipeptide production and microbiome colonization after antibiotic treatment. In vivo mouse studies further showed that lanthipeptide-producing microbes, like Blautia pseudococcoides SCSK, could prevent recovery of microbiome composition and metabolites. Blautia-induced dysbiosis could last for weeks, resulting in prolonged susceptibility to C. difficile and Klebsiella infection. | 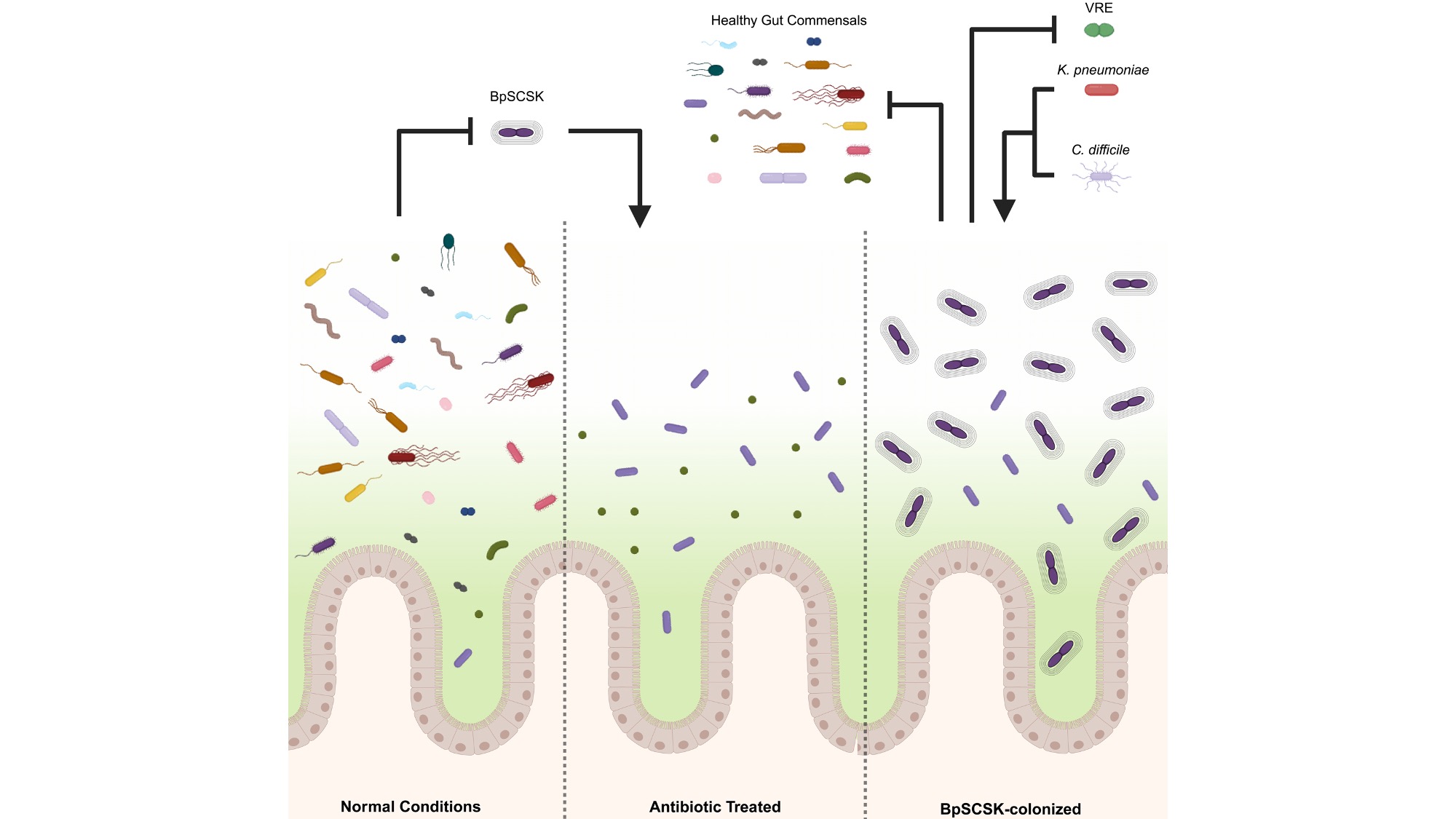 |
| Dinh and Mayuresh, along with collaborators from the van der Donk, Suga, and Goto labs, have published a paper in the Journal of the American Chemical Society defining how peptide aminoacyl-tRNA ligases (PEARLs) recognize their aminoacyl-tRNA substrates. Using flexizyme-generated tRNAs carrying natural and noncanonical amino acids, they mapped substrate preferences of the Bacillus halodurans enzyme BhaBCala. They found that both the acceptor stem and anticodon arm of the tRNA are critical for recognition, while the enzyme tolerates a range of small amino acids, hydroxy acids, and mercaptocarboxylic acids. AlphaFold 3 modeling showed how these RNA elements interface with the active site, enabling future engineering of ligases for forming diverse peptide linkages. |
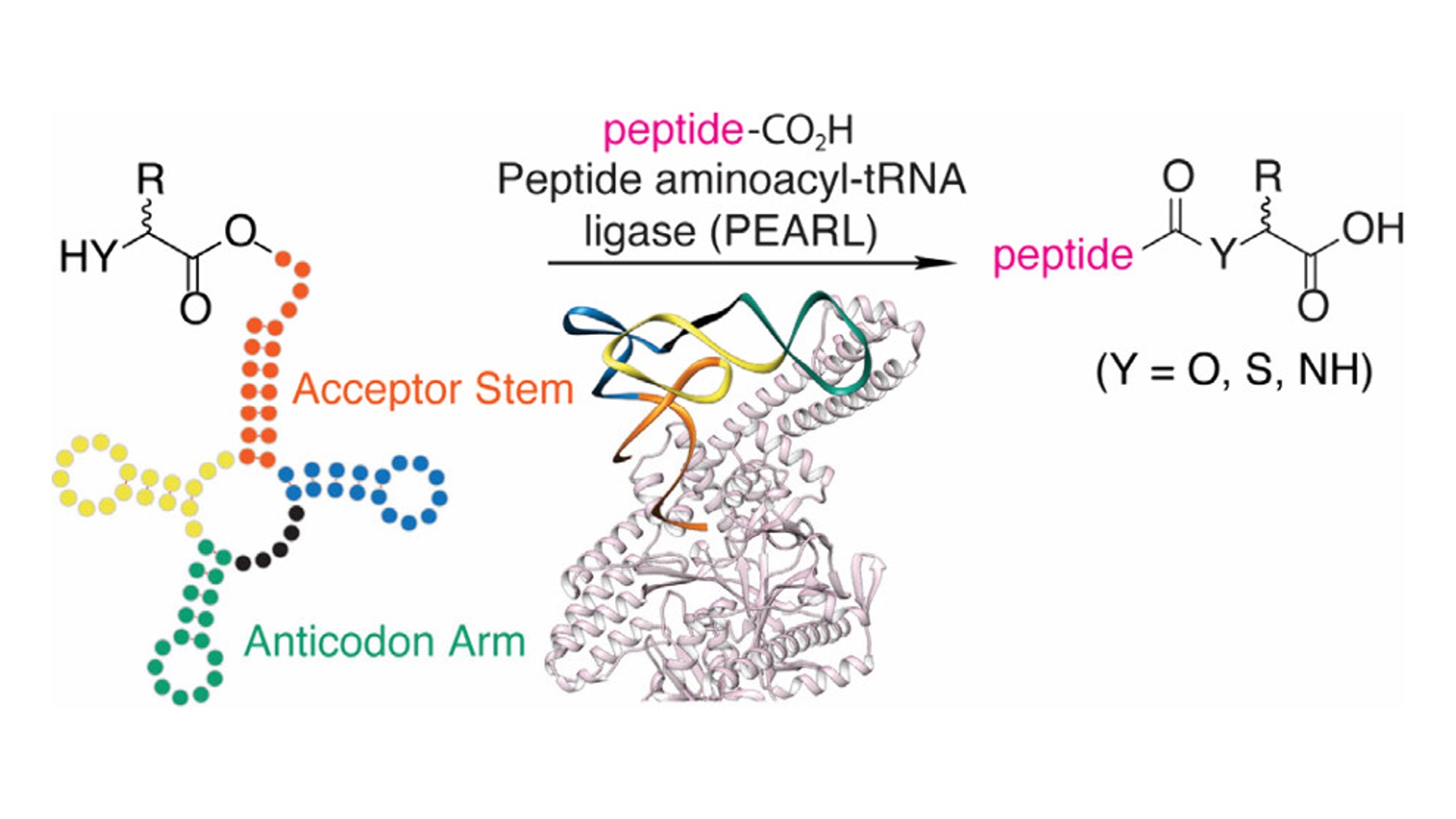 |
| Shravan and collaborators from the Mitchell, Chekan, Zhao, and Sarlah labs have published a paper in ACS Central Science where they explore the biosynthesis and engineerability of daptides. Using in vitro enzyme assays, they elucidate enzymatic requirements for each step in conversion of peptidic C-termini to a tertiary amine group. Local YcaO enzymes additionally convert pathway intermediates to C-terminal imidazoline groups, expanding metabolic diversity within the daptides and the scope of YcaO reactions. Finally, they show the enzymes are remarkably robust, tolerating diverse non-cognate and unnatural peptide sequences. This substrate tolerance allowed them to make unnatural C-terminal analogs and perform bioconjugation on a macrocyclic peptide, the peptide hormone glucagon, and GFP. | 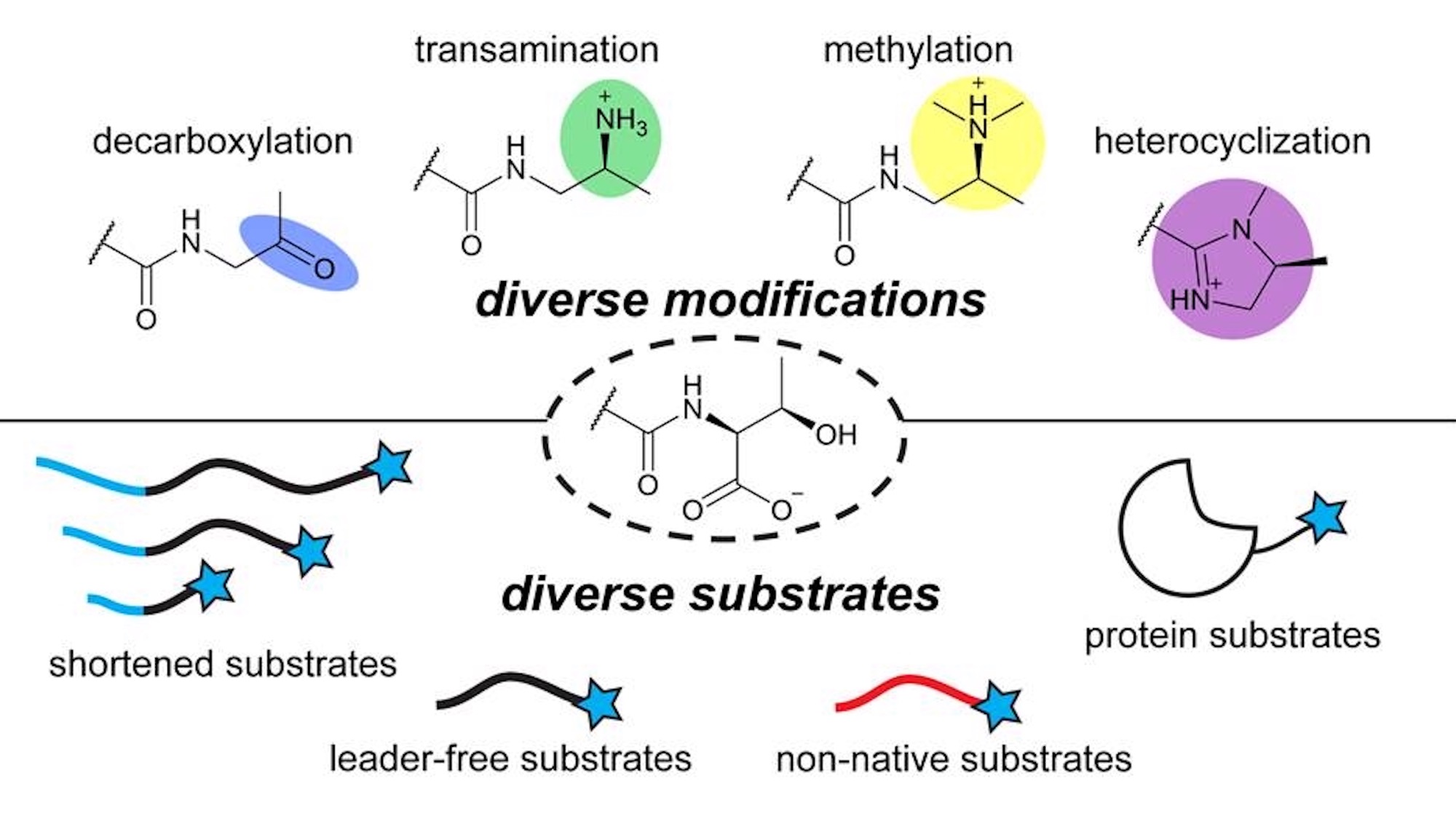 |
| Susanna and collaborators from the Shukla group have published a paper in Nature Communications in which they developed LassoESM, a large language model specifically tailored for lasso peptides. They showed that LassoESM embeddings outperform generic protein language models in predicting lasso cyclase substrate tolerance, in scoring compatibility of non-cognate cyclase-substrate pairs, and even in forecasting RNA polymerase inhibitory activity of lasso peptides. Importantly, the model remains effective under limited training data, and they demonstrated a “human-in-the-loop” framework to iteratively validate and refine predictions. Through this work, they provide a powerful computational tool to accelerate rational design and engineering of functional lasso peptides for biomedical applications. | 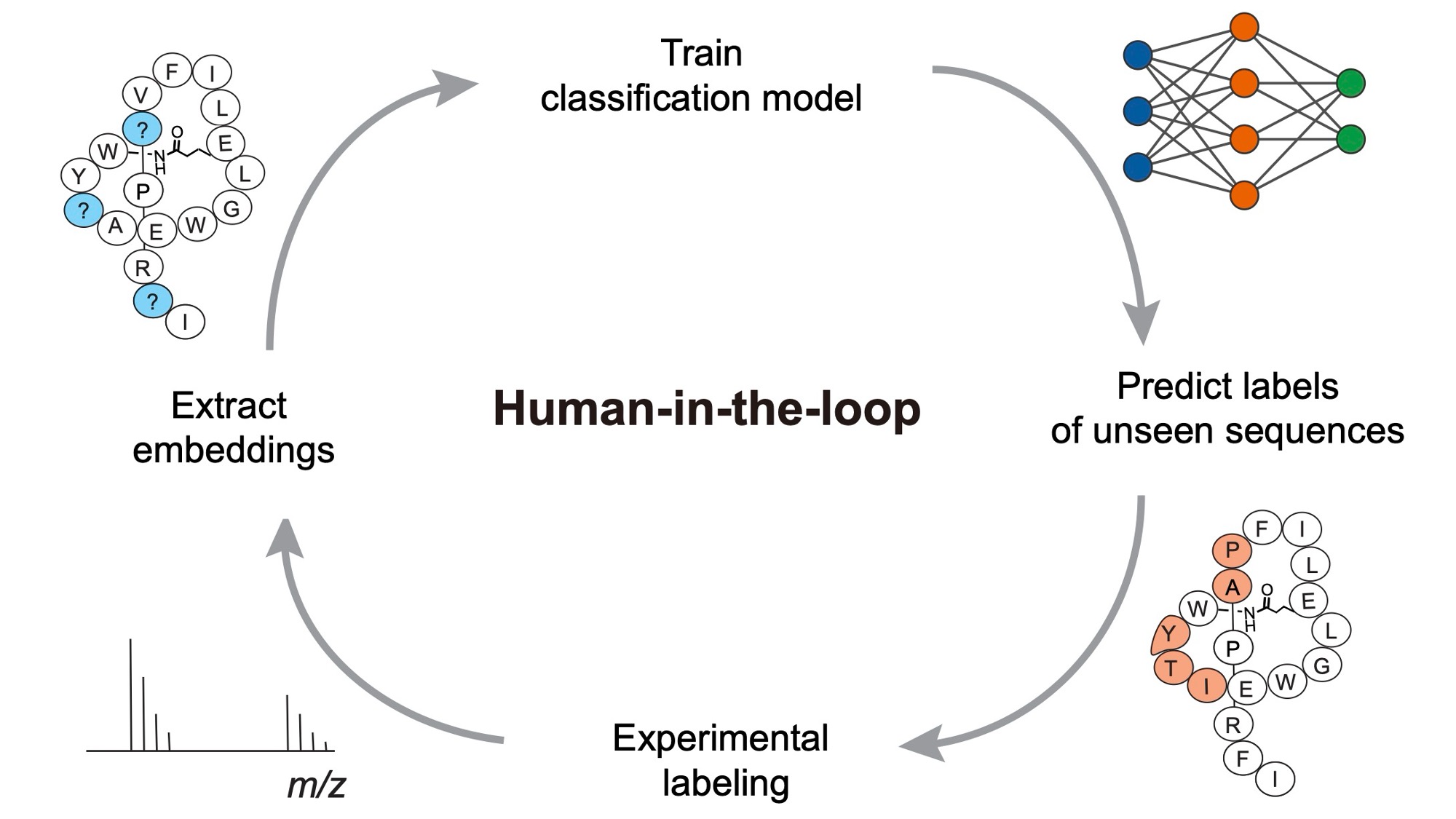 |
| Andrew and Mayuresh, with collaborators from the Mchaourab group, have published a paper in Angewandte Chemie International Edition where they characterized the tryptophan halogenase involved in chlorolassin biosynthesis, ChlH, uncovering remarkably broad substrate promiscuity. ChlH was found to exhibit high selectivity on its native substrate, but modify a diverse array of linear peptides, macrocyclic peptides, and even some proteins in vitro. | 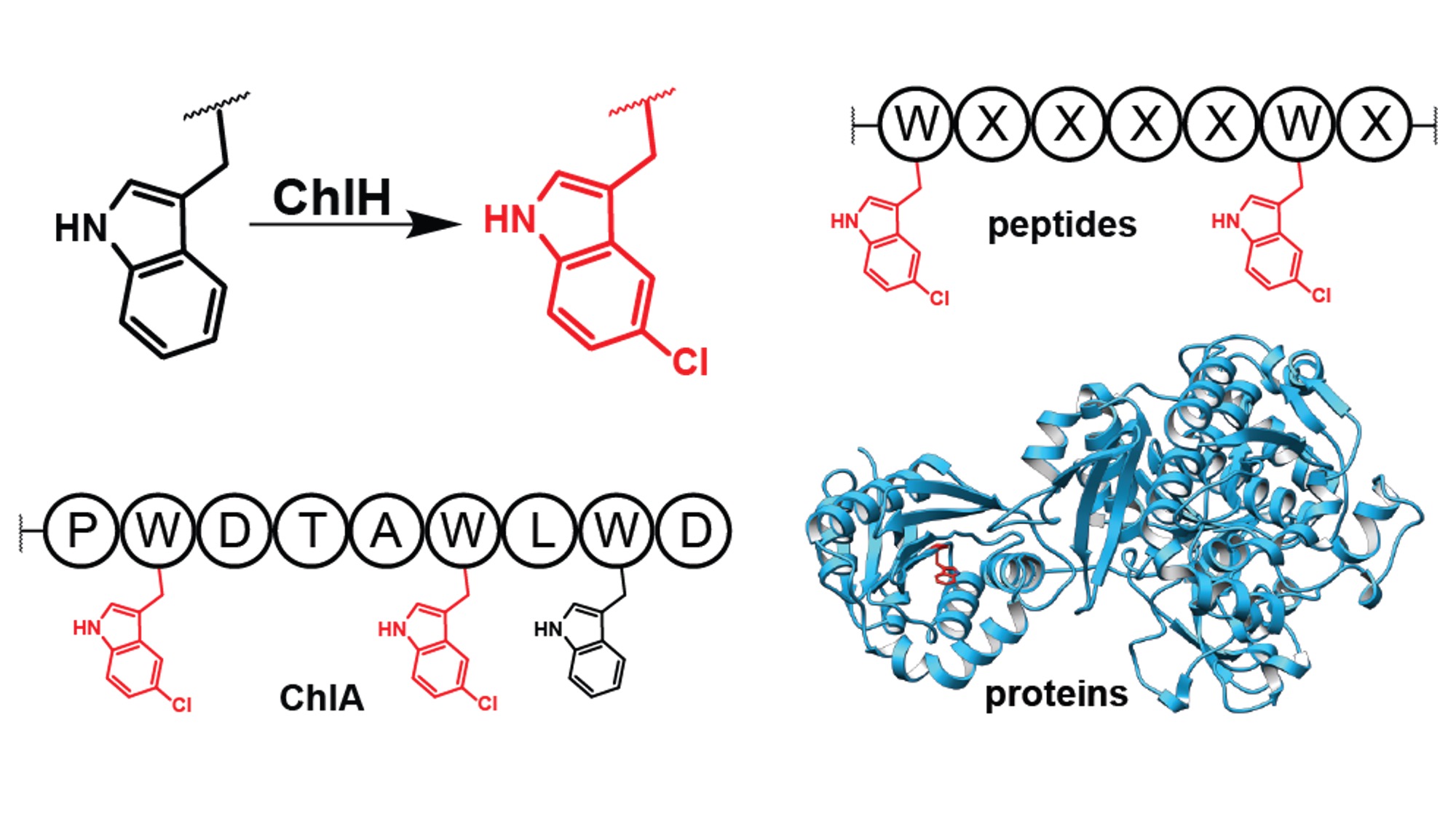 |