Former Projects
In addition to our currently ongoing projects, our lab has had a number of projects that reached satisfying conclusions, or that we have worked on for a time and then moved on from. Here you can find brief summaries of former projects in the lab, along with links to some of the most impactful papers that have come from the projects.
Thiopeptides
We studied the biosynthesis of thiopeptide natural products.
Highlighted Publications
Nguyen, D.T.; Le, T.T.; Rice, A.J.; Hudson, G.A.; van der Donk, W.A.; Mitchell, D.A. “Accessing Diverse Pyridine-Based Macrocyclic Peptides by a Two-Site Recognition Pathway” J. Am. Chem. Soc. (2022). 10.1021/jacs.2c02824
Hudson, G.A.; Hooper, A.R.; DiCaprio, A.J.; Sarlah, D.; Mitchell, D.A. “Structure prediction and synthesis of pyridine-based macrocyclic peptide natural products.” Org. Lett. (2020). doi:10.1021/acs.orglett.0c02699
Schwalen, C.J., Hudson, G.A., et al. “Bioinformatic Expansion and Discovery of Thiopeptide Antibiotics.” J. Am. Chem. Soc. (2018). doi:10.1021/jacs.8b03896
Cogan, D.P., Hudson, G.A., et al. “Structural Insights into Enzymatic [4+2]-Aza-cycloaddition in Thiopeptide Antibiotic Biosynthesis.” Proc. Natl. Acad. Sci. (2017). doi:10.1073/pnas.1716035114
The chemical hallmark of the linear azole-containing peptide (LAP) family of natural products are thiazol(in)e and (methyl)oxazol(in)e heterocycles installed by cyclodehydratases from cysteine and serine/threonine residues, respectively. These post-translational modifications impart rigidity on a peptidic natural product and in many cases are crucial for downstream enzymatic processing and bioactivity. Reported in a series of papers from 2012-2015, we thoroughly investigated the details of cyclodehydration, including the mechanistic enzymology of azoline biosynthesis, which uses ATP to directly activate the peptidic amide backbone, the structural details of the cyclodehydratase, revealing a novel ATP binding motif, and the orchestration of the flavin-dependent dehydrogenase which oxidizes azolines to corresponding azoles. Further research revealed a new class of cyclodehydratases which, upon characterizing, deciphered how the peptidic substrate is recruited to the cyclodehydratase at the molecular level.
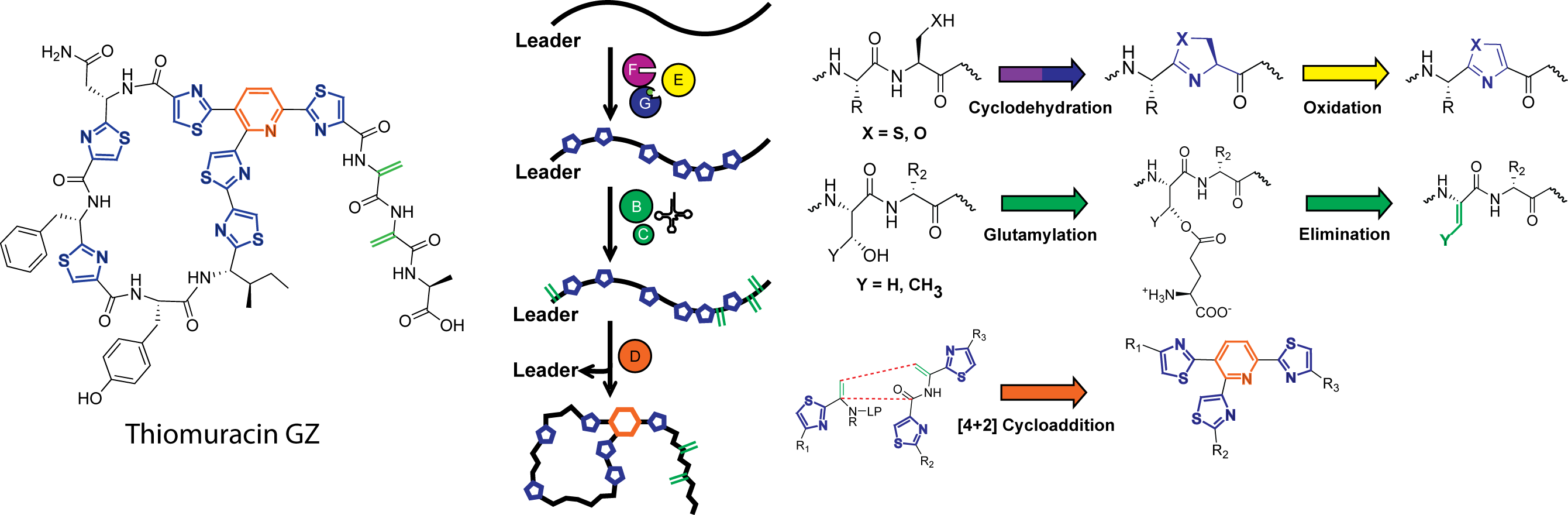
Gaining a deeper understanding of cyclodehydration has enabled the study of more complicated systems, including the thiopeptides, a family of peptidic natural products with potent antibiotic activity towards a variety of drug-resistant bacteria. Despite their architectural complexity, thiopeptide biosynthesis proceeds through a LAP-like intermediate, and as such, feature cyclodehydratases and dehydrogenases for thiazole biosynthesis, but also feature lanthipeptide-like dehydratases and a [4+2] cycloaddition enzyme responsible for pyridine formation. Building upon our previous work on cyclodehydratases, and in collaboration with lanthipeptide biosynthetic experts in the laboratory of Prof. Wilfred van der Donk, we have reconstituted the core biosynthesis of thiomuracin, enabling production of these complicated molecules in vitro using only six enzymes on a precursor peptide substrate. Reconstitution also provided unprecedented insight into the precise details of thiopeptide biosynthesis, including the largely uncharacterized [4+2] cycloaddition enzyme. Further endeavors have yielded crucial insight into the timing and substrate specificity of thiomuracin biosynthesis and will provide a groundwork for enzymatically generating new thiopeptide analogs with desirable pharmacological properties.
Plantazolicin
We characterized the biosynthesis and mode-of-action of this Bacillus anthracis (anthrax) – killing molecule.
Highlighted Publications
Si, T. et al., “Profiling of microbial colonies for high-throughput engineering of multi-step enzymatic reactions via optically guided MALDI MS.” J. Am. Chem. Soc. (2017). doi:10.1021/jacs.7b04641
Deane, C.D. et al., “In vitro biosynthesis and substrate tolerance of the plantazolicin family of natural products.” ACS Chem. Biol. (2016). doi:10.1021/acschembio.6b00369
Molohon, K. et al., “Plantazolicin is an ultra-narrow spectrum antibiotic that targets the Bacillus anthracis membrane.” ACS Infect. Dis. (2016). doi:10.1021/acsinfecdis.5b00115
Hao, Y., Blair, P.M., et al., “Insights into methyltransferase specificity and bioactivity of derivatives of the antibiotic plantazolicin” ACS Chem. Biol. (2015). doi:10.1021/cb501042a
One linear azoline-containing peptide natural product (LAP) that has received a significant amount of our attention is plantazolicin (PZN). This compound is from a soil-dwelling bacterium, Bacillus velezensis (previously classified as Bacillus amyloliquefaciens). In 2008, the biosynthetic gene cluster for PZN was bioinformatically identified; in 2011, we carried out a systematic in vivo dissection of the biosynthetic pathway. In that same year, we determined the chemical structure of PZN using high-resolution MS and multidimensional NMR. This work also revealed an intriguing biological activity for PZN, which displays remarkably selective antibiotic activity against Bacillus anthracis, the causative agent of anthrax. Chemotype-driven bioinformatics methods established that other bacteria produce PZN-like compounds, which now constitute a new structural class of antibiotic.

Studies emerging in 2013, 2016, and 2017 shed light on the structure-activity relationships for the PZN pharmacophore and demonstrated which parts of PZN are necessary for bioactivity and B. anthracis specificity. A detailed chemical and biological investigation into the mode of action implicated cardiolipin-rich microdomains of the B. anthracis plasma membrane as the biological target of PZN.
Anti-virulence
Bacterial pathogens wield an arsenal of virulence factors that permit the establishment and persistence of disease. Taking the logic of the “ultra-narrow” scenario one step further, another strategy for suppressing the rise of antibiotic resistance would be to have a drug that does not kill (nor suppress the growth of) bacteria. Instead, the drug would function via targeting a linchpin virulence factor, rendering the pathogen incapable of causing disease. Relative to conventional antibiotics, an anti-virulence strategy will delay the development of antibiotic resistance by at least two routes: (i) Because disease-causing bacteria employ disparate pathogenic mechanisms, anti-virulence agents
will be inherently ultra-narrow spectrum, negating the evolutionary advantage of laterally transferring resistance cassettes; (ii) Instead of forcing the pathogen to “mutate or die,” it is rendered non-pathogenic and should be cleared from the host like any other bacterium. This reduces the selection of random mutations that diminish drug efficacy.
As attractive as this strategy may seem, it is not without risk or opposition. Critics suggest that it could be dangerous to leave bacteria alive inside the patient. This certainly could be the case with systemic infections, but in general, bacteria, even disease-causing species, are constantly colonizing most parts of the body with no ill effects (only under rare occurrences is a pathogenic attack initiated). Until virulence-targeting drugs are fully evaluated through clinical trials, the utility of such drugs will remain theoretical. We explored antivirulents as a new class of anti-infectives.
Highlighted Publications
Maxson, T., et al., “Targeted treatment for bacterial infections: Prospects for pathogen-specific antibiotics coupled with rapid diagnostics.” Tetrahedron (2015). doi:10.1016/j.tet.2015.09.069
Molloy, E.M., et al., “Identification of the minimal cytolytic unit for streptolysin S and an expansion of the toxin family.” BMC Microbiol. (2015). doi:10.1186/s12866-015-0464-y
Maxson, T., et al., “HIV protease inhibitors block streptolysin S production” ACS Chem. Biol. (2015). doi:10.1021/cb500843r
Zhang, Z., et al., “HIV-1 Integrase Inhibitor-Inspired Antibacterials Targeting Isoprenoid Biosynthesis.” ACS Med. Chem. Lett. (2012). doi:10.1021/ml300038t
Reactivity based screening
We used small-molecule probes to identify natural products with various organic functional groups.
Highlighted Publications
Maxson, T., Tietz, et al., “Targeting reactive carbonyls for identifying natural products and their biosynthetic origins.” J. Am. Chem. Soc., (2016). doi:10.1021/jacs.6b06848
Molloy, E.M., et al., “Biological characterization of the hygrobafilomycin antibiotic JBIR-100 and bioinformatic insights into the hygrolide family of natural products.” Bioorg. Med. Chem., (2016). doi:10.1016/j.bmc.2016.05.021
Cox, C.L., et al., “Nucleophilic 1,4-additions for natural product discovery” ACS Chem. Biol. (2014). doi:10.1021/cb500324n
Genomic sequencing has revealed that Actinobacteria, a historically rich source of FDA-approved drugs, possess far greater natural product biosynthetic potential than previously imagined. One of the challenges associated with discovering novel molecules produced by these unexplored pathways involves determining whether the molecule is being made by the host organism and if the molecule is chemically novel in a process known as dereplication. Rapid determination of chemical novelty is especially of interest since rediscovery of known compounds has led to diminishing returns on investment for the pharmaceutical industry.
Our lab has developed a reactivity-based screening (RBS) strategy to circumvent these long-standing challenges. By taking advantage of chemoselective chemistry, we developed probes to react selectively with functional groups found on various classes of RiPP, polyketide,and non-ribosomal peptide natural products. Upon analysis of differential mass spectrometry data and through database searching, we can rapidly determine the presence and novelty of our target natural products. We took both a bioinformatics-guided as well as comprehensive screening approach with RBS and applied this strategy towards the discovery of thiopeptides, fumarate-containing polyketides as well as aldehyde-containing non-ribosomal peptides.