Discovering Novel Enzyme Chemistry: RiPP Recognition Element
Historically, new RiPPs have been discovered through serendipity. 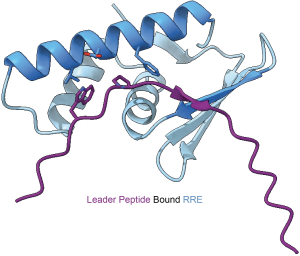 However, the rise of new bioinformatics methods has led us to question whether new RiPP classes can be systematically discovered. As biosynthetic gene clusters (BGCs) for many classes of RiPPs were first reported, we observed a domain commonly involved in leader peptide recognition during biosynthesis. Our current surveys of this domain, the RiPP Recognition Element (RRE), suggest it is encoded in about half of prokaryotic RiPP classes. Despite the lack of sequence similarity between RRE domains, each member shares a common protein fold and employs a similar binding mode to recognize the leader peptide. After our initial report on the RRE domain, we have employed various approaches to study its function in RiPP biosynthesis and leverage its presence to find new enzymatic transformations.
However, the rise of new bioinformatics methods has led us to question whether new RiPP classes can be systematically discovered. As biosynthetic gene clusters (BGCs) for many classes of RiPPs were first reported, we observed a domain commonly involved in leader peptide recognition during biosynthesis. Our current surveys of this domain, the RiPP Recognition Element (RRE), suggest it is encoded in about half of prokaryotic RiPP classes. Despite the lack of sequence similarity between RRE domains, each member shares a common protein fold and employs a similar binding mode to recognize the leader peptide. After our initial report on the RRE domain, we have employed various approaches to study its function in RiPP biosynthesis and leverage its presence to find new enzymatic transformations.
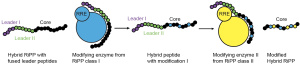
Most of the time, RRE domains require a specific recognition sequence to bind their substrate peptide. We have deployed various biochemical techniques to explore the requirements for RRE domain-precursor peptide interactions across multiple RiPP classes. Using this information, we have engineered new-to-nature molecules by grafting different recognition sequences into the same peptide sequence. Our in-depth mechanistic investigations have also demonstrated how the RRE domain is key in the enzymatic activation of RiPP-modifying enzymes.
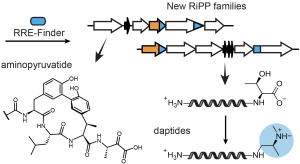
Given the widespread nature of RRE domains, we hypothesized that predicting RRE domains could enable the discovery of new RiPP pathways. We developed a tool, RRE-Finder, to rapidly survey RRE domains across different genomes. Using our curated dataset of thousands of RRE domains, we have searched genomic data for new BGCs with RRE domains, and we have identified numerous new classes of RiPPs through these efforts. We have already published two of these studies discovering the daptides, a class of peptides with two amino termini, and aminopyruvatide, a RiPP modified by three different metalloenzymes. Further efforts are underway to expand known enzyme chemistry using RRE-domain-based discovery and explore the biological functions of these newly identified pathways.
Key Publications: