Characterizing New Enzymes: YcaO Enzymes
When asked for a post-translational modification, most cite a sidechain modification since we are taught as undergraduates that mainchain amides are relatively unreactive. However, RiPPs illustrate that Nature exploits the mainchain in many creative ways. In the lab’s early days, we embarked on a campaign to investigate enzymatic modifications to peptide backbones. We uncovered an unusual, ATP-dependent cyclodehydration mechanism for “YcaO” enzymes. YcaO cyclodehydratases transform nucleophilic (Cys/Ser/Thr) residues by facilitating attack on the preceding amide and phosphorylating the backbone oxygen prior to yielding thiazoline/oxazoline heterocycles. Formally, YcaO acts as a mainchain-modifying kinase. We systematically characterized several YcaOs to reveal residues involved in substrate binding, catalysis, structural integrity, and partner protein affinity. With Satish Nair, we reported several crystal structures of a YcaO from E.coli, which presents a new protein fold. These revealed an unprecedented ATP binding motif. The E. coli YcaO enzyme was devoid of cyclodehydration activity and lacked a locally encoded precursor peptide, which prompted an investigation into possible other YcaO catalyzed reactions.
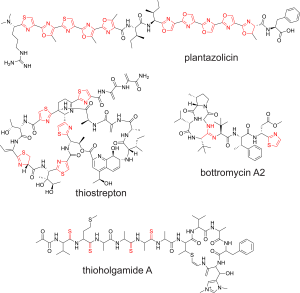
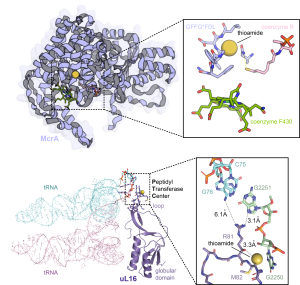
Utilization of additional, and sometimes exogenous, nucleophiles expand the potential chemistries catalyzed by YcaOs. Indeed, amines can form macrolactamidines, exemplified by the antibiotic bottromycin, while sulfides form thioamides, exemplified by the antiproliferative thioholgamide. Thioamide-forming YcaOs caught our attention for both their rarity and simplicity: a mainchain oxygen/sulfur swap drastically alters backbone conformation, protease resistance, and hydrogen-bonding, despite constituting only a single-atom change. While prevalent in RiPPs, their presence in proteins is exceedingly rare. At the time, methyl-coenzyme M reductase (MCR), which produces methane and is a key player in the global carbon cycle, was the only known thioamidated protein. Using a model methanogen, we worked with Bill Metcalf to show that YcaO and TfuA (an adjacent DUF) were necessary and sufficient for MCR thioamidation in vivo and in vitro. Fitness correlated with thioamidation, as mutants lacking thioamidation divided 50% slower than wildtype. Furthermore, thioamidation is critical for the organism’s survival at elevated temperatures. These are profound effects for a single atom replacement. We further elucidated the role of TfuA, which is part of a sulfide-delivery mechanism for thioamide-forming YcaOs. TfuA is a formal thiocarboxylate hydrolase, which liberates H2S from the C-terminus of a carrier protein, and an allosteric activator of YcaO. Beyond methanogens, the abovementioned E. coli YcaO has been shown to perform protein thioamidation of ribosomal protein uL16. The modification site is proximal to the peptidyltransferase center and may influence translational rate/fidelity.
Our bioinformatic surveys have identified ~20,000 YcaOs, many of which remain uncharacterized. The functional assignment of and exploration of biological roles for YcaO enzymes remains an active and thriving area of investigation in the lab.
Key Publications:
4. Mechanistic basis for ribosomal peptide backbone modifications. ACS Cent. Sci., 5: 842-851 (2019).