Research
Research Description
How do you organize the crowded environment of a cell? Work over the past decade has shown that the cytosol and nucleoplasm are a non-uniform heterogeneous mixture organized by membrane-less organelles called biomolecular condensates. Unlike traditional organelles, such as the nucleus and mitochondria which have lipid membranes to facilitate the physical compartmentalization of cellular components, biomolecular condensates lack such physical barriers. Rather, compartmentalization is facilitated by collective interactions of biomolecules which form highly concentrated assemblies and, in some cases, resemble a spherical puncta within the cell.
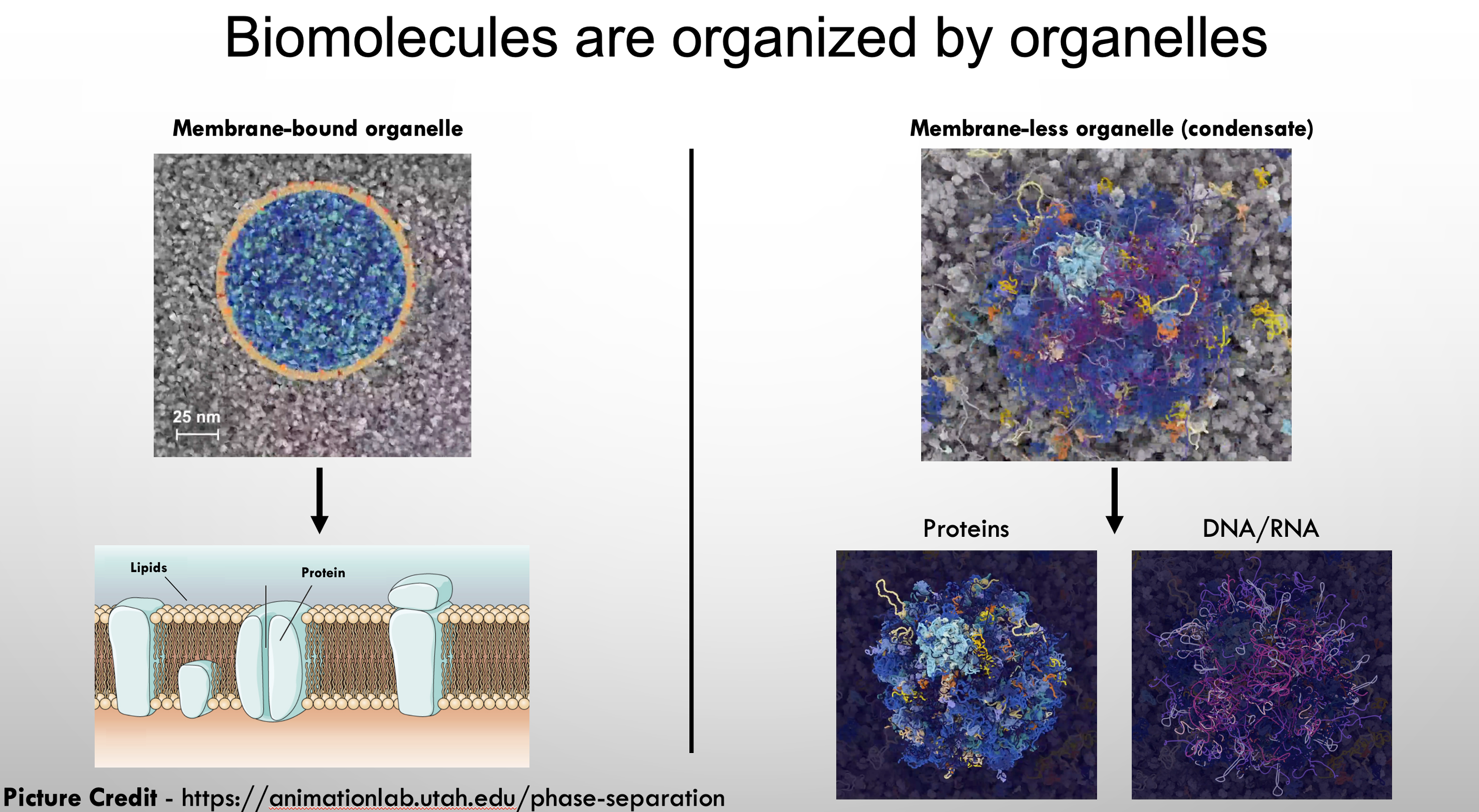
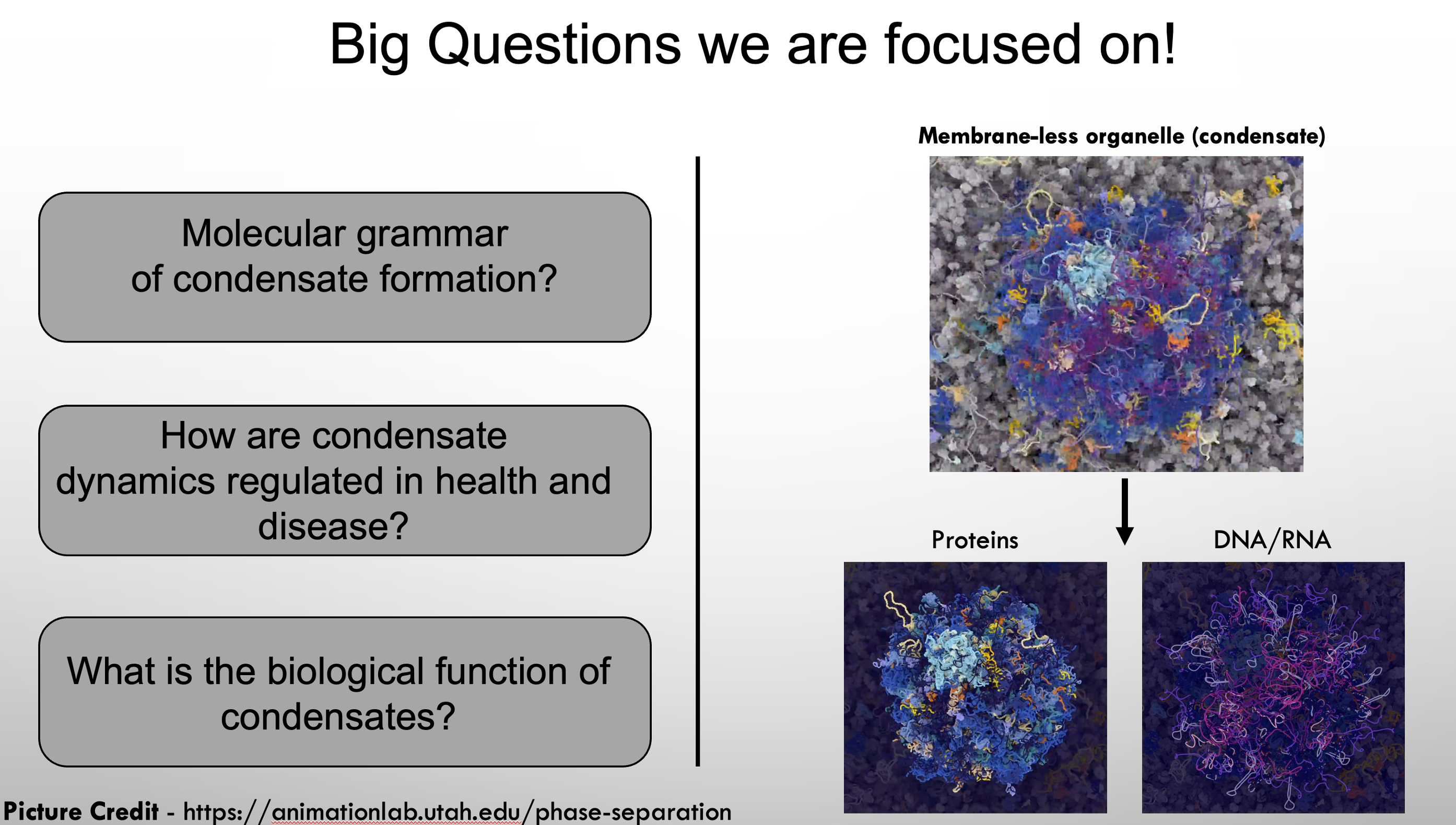
Biomolecular condensates are dynamic, highly regulated, and sensitive to environmental changes, including the cell cycle, post-translational modifications, and stress. Condensates contain both proteins and nucleic acids and have been linked to many biological processes. However, in most processes the functional role of condensates in living cells has not been experimentally demonstrated. To investigate condensates in living cells, we utilize several model systems including the nematode worm C. elegans, tissue culture, and in vitro reconstitution. Using these condensate models my laboratory is interested in understanding the molecular mechanisms that drive condensate function and regulation.
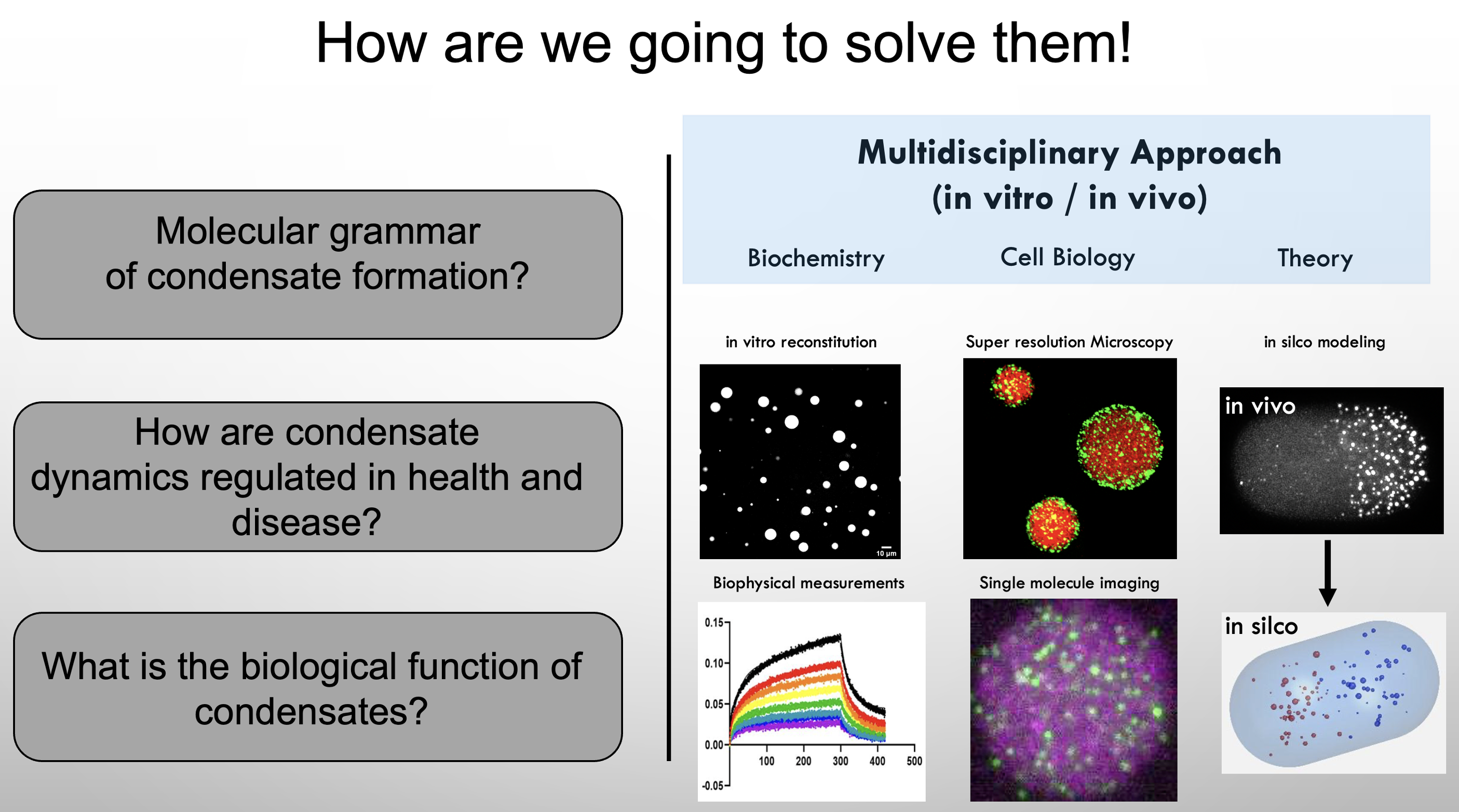
To gain fundamental mechanistic insight into condensate function and regulation we utilize a diverse array of state-of-the-art approaches including CRISPR genome editing, in vitro reconstitution, biophysical measurements, single-molecule microscopy, super-resolution microscopy, and in silco modeling. The long-term goal of my laboratory is to understand the integration of biomolecular condensate regulation and function across biological time scales: from molecules – to cells – through animal development.