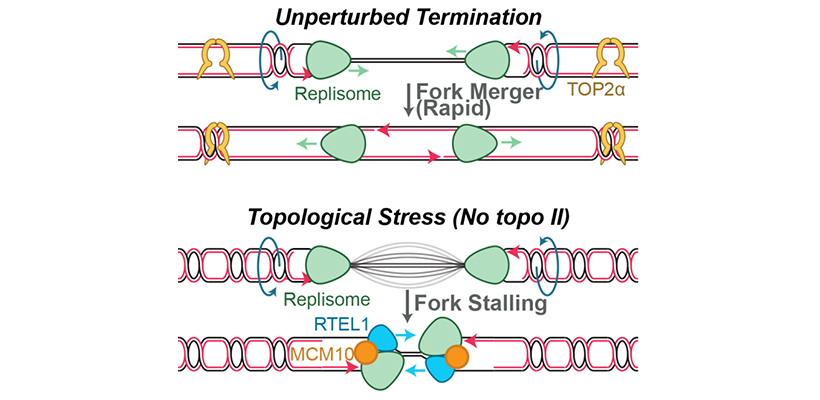
Endogenous and exogenous genotoxins can cause up to ~10,000 replication forks to stall during each S phase. In response, the replication fork junction is regressed and nascent DNA strands are extruded (‘fork reversal’). Nascent DNA strands are also subject to degradation by nucleases (‘nascent strand degradation’). Fork reversal and nascent strand degradation are thought to allow replication forks to overcome DNA lesions and restart DNA synthesis, in order to promote genome stability. However, fork reversal and nascent strand degradation can also cause genome instability when they occur excessively. It is thus important to understand how these processes are triggered efficiently, but not spuriously. Additionally, fork reversal and nascent strand degradation involve many proteins that are essential for viability and/or implicated in disease. Understanding these processes is therefore crucial to understanding how these important proteins function.
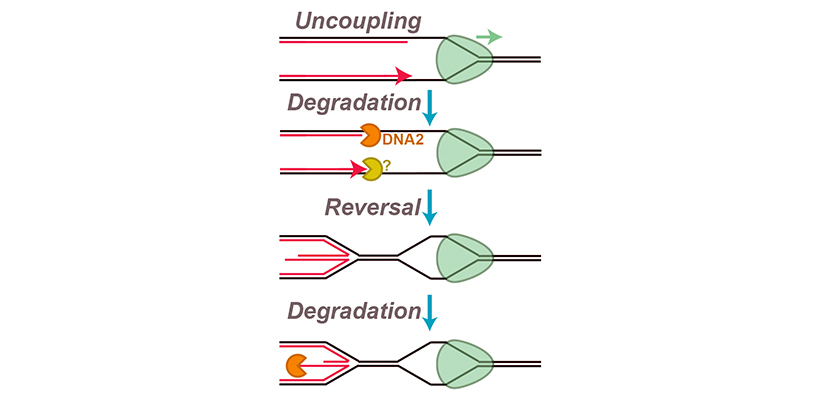
We are interested in trying to understand how fork reversal and nascent strand degradation are triggered and how exactly the underlying molecular events unfold. We recently developed a new in vitro approach to study nascent strand degradation and fork reversal, which we corroborated with cellular experiments. Surprisingly, we found that a subset of events involved in fork stalling are sufficient to cause fork reversal and nascent strand degradation. Specifically, we showed that these events can be triggered by replication fork ‘uncoupling’, which occurs when the DNA polymerases stall but the replication fork helicase keeps unwinding. We also identified an additional degradation step, prior to fork reversal, and found that the replicative helicase is retained throughout this process. We are now studying exactly how uncoupling triggers these events, and how different proteins participate in fork reversal and nascent strand degradation.
Health relevance: Fork reversal and nascent strand degradation are a primary response to genotoxins derived from either the environment (e.g. UV and ionizing radiation) or natural cellular metabolism (e.g. reactive oxygen species). It is important to understand how these processes are triggered and how they work so that we can better evaluate the risk of exposure to genotoxins. Fork reversal and nascent strand degradation involve tumor suppressors (e.g. BRCA1, BRCA2). Studying the role of these proteins during fork reversal and nascent strand degradation will allow us to better understand how they mitigate cancer development. Finally, defects in replication restart are targeted during cancer chemotherapy so understanding these pathways should inform the development of improved cancer chemotherapies.