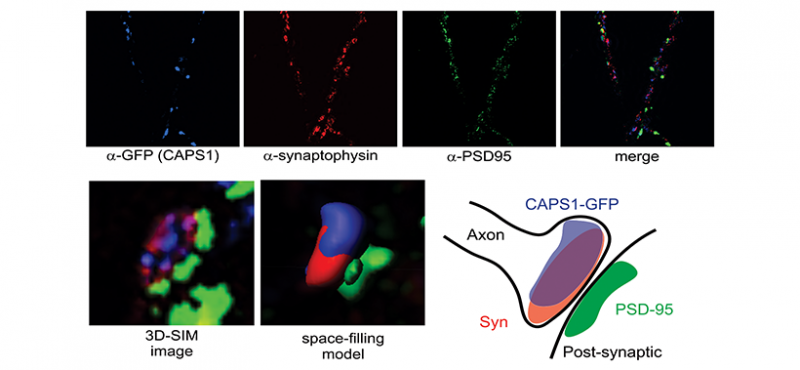
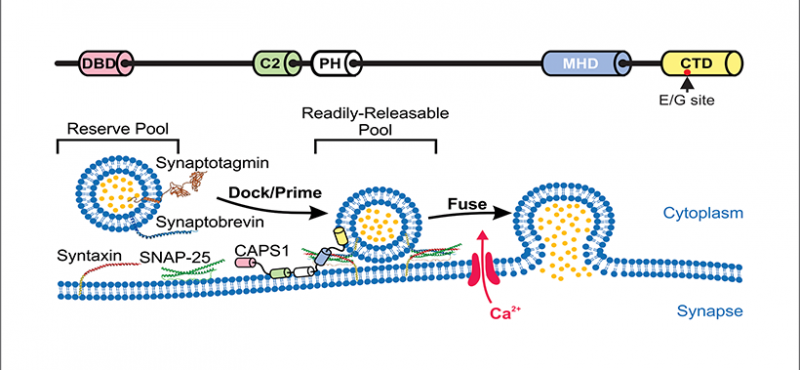
Welcome to the Emeson Lab
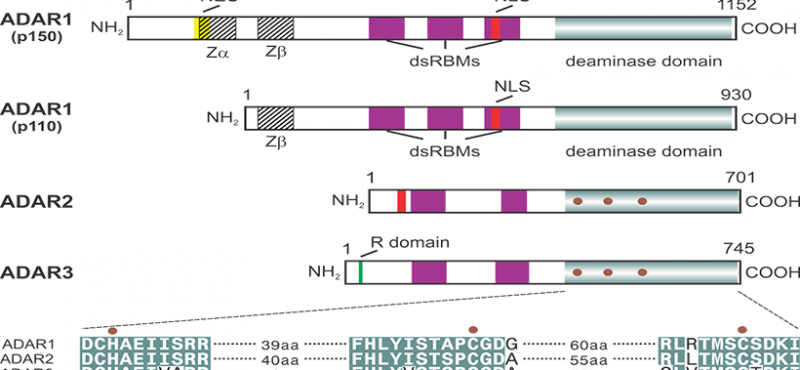
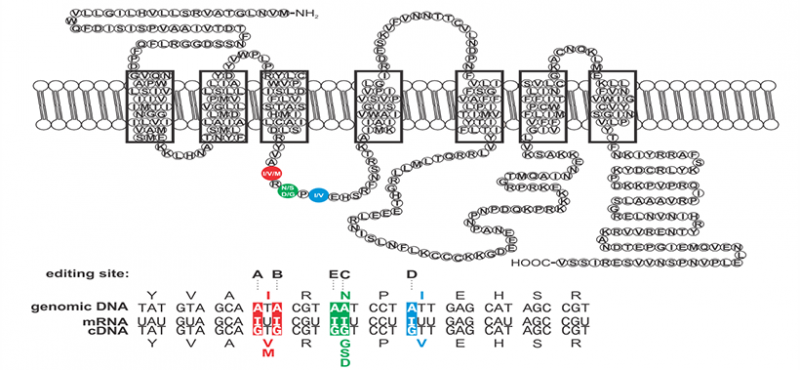
The research interests of our laboratory focus on the cellular and molecular processes underlying neuronal communication in normal and pathophysiologic disease states. Specifically, we are examining the molecular mechanisms involved in the editing of RNA transcripts encoding proteins critical for mammalian nervous system function. RNA editing is a post-transcriptional modification in which specific adenosine residues in pre-messenger RNAs are converted to inosine (A-to-I editing) by double-stranded RNA-specific adenosine deaminases (ADARs). As a result of these deamination events, the coding potential of RNAs can be subtly altered to change as little as a single amino acid residue in resultant products to generate protein isoforms with distinct functional properties.
Programmable alterations of nucleic acid sequence offer significant therapeutic potential for a wide range of genetic disorders. In recent years, the RNA molecule has become one of the most promising targets for therapeutic intervention and many RNA-based therapeutics have been developed recently. Despite the promise of CRISPR/Cas9-based approaches for the editing of genomic DNA, this paradigm has numerous disadvantages, including off-target and imprecise modifications, as well as a low efficiency rate in post-mitotic cell types such as neurons.
Our current research efforts focus upon an alternative approach for targeted genome engineering using cellular processes normally involved in the A-to-I editing of RNA transcripts. These studies involve the development of a systematic pipeline for the selection of ADAR-recruiting RNAs (arRNAs) that will promote efficient, site-selective adenosine deamination to repair common mutations associated with neurological disorders such as Parkinson’s Disease and Rett Syndrome.
It is anticipated that these studies will allow the pre-clinical development of therapeutic arRNAs for the repair of numerous genomic variants leading to disease in humanized mouse models and affected patients.